Human Coronavirus OC43 as a Low-Risk Model to Study COVID-19
Abstract
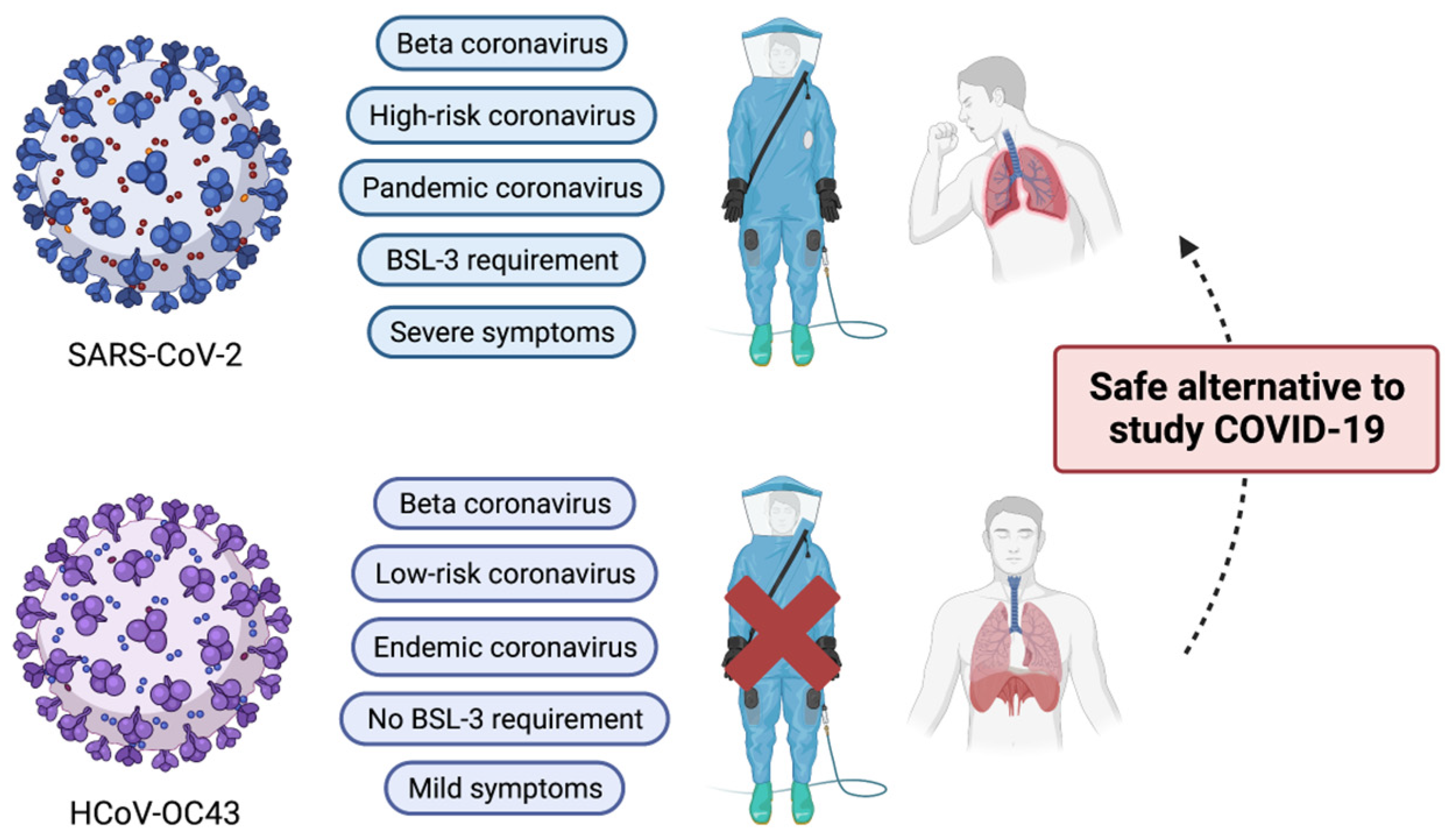
:1. Introduction
2. Classification of Human Coronaviruses
3. Brief History of HCoV-OC43
4. Clinical Manifestations of HCoV-OC43 Infection
5. Genome Structure of HCoV-OC43
6. Characteristics of Structural Proteins of HCoV-OC43
6.1. Hemagglutinin-Esterase
6.2. Spike Protein
6.3. Envelope Protein
6.4. Membrane Protein
6.5. Nucleocapsid
7. Evolution of HCoV-OC43
8. Host Interactions with HCoV-OC43
8.1. Virus Entry Factors
8.2. Intracellular Host Factors
8.3. Host Immunological Responses
9. Neuropathology by HCoV-OC43 Infection
9.1. In Vitro Model
9.2. In Vivo Model
9.3. Clinical Model
10. Discovery of Antiviral Candidates by Using HCoV-OC43
11. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Masters, P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006, 66, 193–292. [Google Scholar] [CrossRef]
- Cimolai, N. Complicating Infections Associated with Common Endemic Human Respiratory Coronaviruses. Health Secur. 2020, 19, 195–208. [Google Scholar] [CrossRef]
- Gaunt, E.R.; Hardie, A.; Claas, E.C.; Simmonds, P.; Templeton, K.E. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J. Clin. Microbiol. 2010, 48, 2940–2947. [Google Scholar] [CrossRef] [Green Version]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graham, N.M. The epidemiology of acute respiratory infections in children and adults: A global perspective. Epidemiol. Rev. 1990, 12, 149–178. [Google Scholar] [CrossRef] [PubMed]
- Hulswit, R.J.G.; Lang, Y.; Bakkers, M.J.G.; Li, W.; Li, Z.; Schouten, A.; Ophorst, B.; van Kuppeveld, F.J.M.; Boons, G.J.; Bosch, B.J.; et al. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc. Natl. Acad. Sci. USA 2019, 116, 2681–2690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Chen, X.; Hu, T.; Li, J.; Song, H.; Liu, Y.; Wang, P.; Liu, D.; Yang, J.; Holmes, E.C.; et al. A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein. Curr. Biol. 2020, 30, 2196–2203.e2193. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prince, T.; Smith, S.L.; Radford, A.D.; Solomon, T.; Hughes, G.L.; Patterson, E.I. SARS-CoV-2 Infections in Animals: Reservoirs for Reverse Zoonosis and Models for Study. Viruses 2021, 13, 494. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.L.; Yeh, I.J.; Phan, N.N.; Wu, Y.H.; Yen, M.C.; Hung, J.H.; Chiao, C.C.; Chen, C.F.; Sun, Z.; Jiang, J.Z.; et al. Gene signatuRes. of SARS-CoV/SARS-CoV-2-infected ferret lungs in short- and long-term models. Infect. Genet. Evol. 2020, 85, 104438. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, K.; Dees, J.H.; Becker, W.B.; Kapikian, A.Z.; Chanock, R.M. Recovery in tracheal organ cultuRes. of novel viruses from patients with respiratory disease. Proc. Natl. Acad. Sci. USA 1967, 57, 933–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahn, J.S.; McIntosh, K. History and recent advances in coronavirus discovery. Pediatr. Infect. Dis. J. 2005, 24, S223–S227, discussion S226. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, K.; Becker, W.B.; Chanock, R.M. Growth in suckling-mouse brain of “IBV-like” viruses from patients with upper respiratory tract disease. Proc. Natl. Acad. Sci. USA 1967, 58, 2268–2273. [Google Scholar] [CrossRef] [Green Version]
- Bracci, N.; Pan, H.C.; Lehman, C.; Kehn-Hall, K.; Lin, S.C. Improved plaque assay for human coronaviruses 229E and OC43. PeerJ. 2020, 8, e10639. [Google Scholar] [CrossRef]
- Kaye, H.S.; Dowdle, W.R. Some characteristics of hemagglutination of certain strains of “IBV-like” virus. J. Infect. Dis. 1969, 120, 576–581. [Google Scholar] [CrossRef] [PubMed]
- Myint, S.; Johnston, S.; Sanderson, G.; Simpson, H. Evaluation of nested polymerase chain methods for the detection of human coronaviruses 229E and OC43. Mol. Cell. Probes 1994, 8, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Wege, H.; Siddell, S.; ter Meulen, V. The biology and pathogenesis of coronaviruses. Curr. Top. Microbiol. Immunol. 1982, 99, 165–200. [Google Scholar] [CrossRef] [Green Version]
- Collins, A.R. Human coronavirus OC43 interacts with major histocompatibility complex class I molecules at the cell surface to establish infection. Immunol. Investig. 1994, 23, 313–321. [Google Scholar] [CrossRef] [Green Version]
- Ogimi, C.; Kim, Y.J.; Martin, E.T.; Huh, H.J.; Chiu, C.H.; Englund, J.A. What’s New With the Old Coronaviruses? J. Pediatr. Infect. Dis. Soc. 2020, 9, 210–217. [Google Scholar] [CrossRef] [Green Version]
- Jean, A.; Quach, C.; Yung, A.; Semret, M. Severity and outcome associated with human coronavirus OC43 infections among children. Pediatr. Infect. Dis. J. 2013, 32, 325–329. [Google Scholar] [CrossRef]
- Kim, J.M.; Jeon, J.S.; Kim, J.K. Climate and Human coronaviruses 229E and Human coronaviruses OC43 Infections: Respiratory Viral Infections Prevalence in Hospitalized Children in Cheonan, Korea. J. Microbiol. Biotechnol. 2020, 30, 1495–1499. [Google Scholar] [CrossRef]
- Talbot, H.K.; Shepherd, B.E.; Crowe, J.E., Jr.; Griffin, M.R.; Edwards, K.M.; Podsiad, A.B.; Tollefson, S.J.; Wright, P.F.; Williams, J.V. The pediatric burden of human coronaviruses evaluated for twenty years. Pediatr. Infect. Dis. J. 2009, 28, 682–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, S.K.; Lee, P.; Tsang, A.K.; Yip, C.C.; Tse, H.; Lee, R.A.; So, L.Y.; Lau, Y.L.; Chan, K.H.; Woo, P.C.; et al. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J. Virol. 2011, 85, 11325–11337. [Google Scholar] [CrossRef] [Green Version]
- Monto, A.S.; Lim, S.K. The Tecumseh study of respiratory illness. VI. Frequency of and relationship between outbreaks of coronavirus infection. J. Infect. Dis. 1974, 129, 271–276. [Google Scholar] [CrossRef] [Green Version]
- Morgello, S. Coronaviruses and the central nervous system. J. Neurovirol. 2020, 26, 459–473. [Google Scholar] [CrossRef] [PubMed]
- Nickbakhsh, S.; Ho, A.; Marques, D.F.P.; McMenamin, J.; Gunson, R.N.; Murcia, P.R. Epidemiology of Seasonal Coronaviruses: Establishing the Context for the Emergence of Coronavirus Disease 2019. J. Infect. Dis. 2020, 222, 17–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saib, I.; Aleisa, S.; Ardah, H.; Mahmoud, E.; Alharbi, A.O.; Alsaedy, A.; Aljohani, S.; Alshehri, A.; Alharbi, N.K.; Bosaeed, M. Non-SARS Non-MERS Human Coronaviruses: Clinical Characteristics and Outcome. Pathogens 2021, 10, 1549. [Google Scholar] [CrossRef]
- Vijgen, L.; Keyaerts, E.; Moes, E.; Thoelen, I.; Wollants, E.; Lemey, P.; Vandamme, A.M.; Van Ranst, M. Complete genomic sequence of human coronavirus OC43: Molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 2005, 79, 1595–1604. [Google Scholar] [CrossRef] [Green Version]
- El-Sahly, H.M.; Atmar, R.L.; Glezen, W.P.; Greenberg, S.B. Spectrum of clinical illness in hospitalized patients with “common cold” virus infections. Clin. Infect. Dis. 2000, 31, 96–100. [Google Scholar] [CrossRef] [Green Version]
- Gagneur, A.; Sizun, J.; Vallet, S.; Legr, M.C.; Picard, B.; Talbot, P.J. Coronavirus-related nosocomial viral respiratory infections in a neonatal and paediatric intensive care unit: A prospective study. J. Hosp. Infect. 2002, 51, 59–64. [Google Scholar] [CrossRef]
- St-Jean, J.R.; Jacomy, H.; Desforges, M.; Vabret, A.; Freymuth, F.; Talbot, P.J. Human respiratory coronavirus OC43: Genetic stability and neuroinvasion. J. Virol. 2004, 78, 8824–8834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, J.N.; Mounir, S.; Talbot, P.J. Human coronavirus gene expression in the brains of multiple sclerosis patients. Virology 1992, 191, 502–505. [Google Scholar] [CrossRef] [PubMed]
- Murray, R.S.; Brown, B.; Brian, D.; Cabirac, G.F. Detection of coronavirus RNA and antigen in multiple sclerosis brain. Ann. Neurol. 1992, 31, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Rajapakse, N.; Dixit, D. Human and novel coronavirus infections in children: A review. Paediatr. Int. Child. Health 2020, 41, 36–55. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Quan, Y.; Xin, Z.T.; Wrammert, J.; Ma, M.J.; Lv, H.; Wang, T.B.; Yang, H.; Richardus, J.H.; Liu, W.; et al. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: A six-year follow-up study. J. Immunol. 2011, 186, 7264–7268. [Google Scholar] [CrossRef] [Green Version]
- Leao, J.C.; Gusmao, T.P.L.; Zarzar, A.M.; Leao Filho, J.C.; Barkokebas Santos de Faria, A.; Morais Silva, I.H.; Gueiros, L.A.M.; Robinson, N.A.; Porter, S.; Carvalho, A.A.T. Coronaviridae-Old friends, new enemy! Oral Dis. 2020, 28, 858–866. [Google Scholar] [CrossRef]
- Marra, M.A.; Jones, S.J.; Astell, C.R.; Holt, R.A.; Brooks-Wilson, A.; Butterfield, Y.S.; Khattra, J.; Asano, J.K.; Barber, S.A.; Chan, S.Y.; et al. The Genome sequence of the SARS-associated coronavirus. Science 2003, 300, 1399–1404. [Google Scholar] [CrossRef] [Green Version]
- Labonte, P.; Mounir, S.; Talbot, P.J. Sequence and expression of the ns2 protein gene of human coronavirus OC43. J. Gen. Virol. 1995, 76 Pt 2, 431–435. [Google Scholar] [CrossRef]
- Forni, D.; Cagliani, R.; Clerici, M.; Sironi, M. Molecular Evolution of Human Coronavirus Genomes. Trends Microbiol. 2017, 25, 35–48. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Ma, Z.; Chen, H.; Lu, Y.; Chen, X. Valinomycin as a potential antiviral agent against coronaviruses: A review. Biomed. J. 2020, 43, 414–423. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J.; Xiao, Y.; Zhang, J.; Wang, Y.; Chen, L.; Paranhos-Baccala, G.; Ren, L.; Wang, J. Genotype shift in human coronavirus OC43 and emergence of a novel genotype by natural recombination. J. Infect. 2015, 70, 641–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beidas, M.; Chehadeh, W. PCR array profiling of antiviral genes in human embryonic kidney cells expressing human coronavirus OC43 structural and accessory proteins. Arch. Virol. 2018, 163, 2065–2072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beidas, M.; Chehadeh, W. Effect of Human Coronavirus OC43 Structural and Accessory Proteins on the Transcriptional Activation of Antiviral Response Elements. Intervirology 2018, 61, 30–35. [Google Scholar] [CrossRef]
- Dolliver, S.M.; Kleer, M.; Bui-Marinos, M.P.; Ying, S.; Corcoran, J.A.; Khaperskyy, D.A. Nsp1 proteins of human coronaviruses HCoV-OC43 and SARS-CoV2 inhibit stress granule formation. PLoS Pathog. 2022, 18, e1011041. [Google Scholar] [CrossRef] [PubMed]
- Mounir, S.; Labonte, P.; Talbot, P.J. Characterization of the nonstructural and spike proteins of the human respiratory coronavirus OC43: Comparison with bovine enteric coronavirus. Adv. Exp. Med. Biol. 1993, 342, 61–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, M.M.; Cavanagh, D. The molecular biology of coronaviruses. Adv. Virus Res. 1997, 48, 1–100. [Google Scholar]
- Zhang, X.M.; Kousoulas, K.G.; Storz, J. The hemagglutinin/esterase gene of human coronavirus strain OC43: Phylogenetic relationships to bovine and murine coronaviruses and influenza C virus. Virology 1992, 186, 318–323. [Google Scholar] [CrossRef]
- Luytjes, W.; Bredenbeek, P.J.; Noten, A.F.; Horzinek, M.C.; Spaan, W.J. Sequence of mouse hepatitis virus A59 mRNA 2: Indications for RNA recombination between coronaviruses and influenza C virus. Virology 1988, 166, 415–422. [Google Scholar] [CrossRef]
- Lissenberg, A.; Vrolijk, M.M.; van Vliet, A.L.; Langereis, M.A.; de Groot-Mijnes, J.D.; Rottier, P.J.; de Groot, R.J. Luxury at a cost? Recombinant mouse hepatitis viruses expressing the accessory hemagglutinin esterase protein display reduced fitness in vitro. J. Virol. 2005, 79, 15054–15063. [Google Scholar] [CrossRef] [Green Version]
- Kienzle, T.E.; Abraham, S.; Hogue, B.G.; Brian, D.A. Structure and orientation of expressed bovine coronavirus hemagglutinin-esterase protein. J. Virol. 1990, 64, 1834–1838. [Google Scholar] [CrossRef] [Green Version]
- King, B.; Brian, D.A. Bovine coronavirus structural proteins. J. Virol. 1982, 42, 700–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, B.; Potts, B.J.; Brian, D.A. Bovine coronavirus hemagglutinin protein. Virus Res. 1985, 2, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Vlasak, R.; Luytjes, W.; Leider, J.; Spaan, W.; Palese, P. The E3 protein of bovine coronavirus is a receptor-destroying enzyme with acetylesterase activity. J. Virol. 1988, 62, 4686–4690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schultze, B.; Wahn, K.; Klenk, H.D.; Herrler, G. Isolated HE-protein from hemagglutinating encephalomyelitis virus and bovine coronavirus has receptor-destroying and receptor-binding activity. Virology 1991, 180, 221–228. [Google Scholar] [CrossRef]
- Zeng, Q.; Langereis, M.A.; van Vliet, A.L.; Huizinga, E.G.; de Groot, R.J. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 9065–9069. [Google Scholar] [CrossRef] [Green Version]
- de Groot, R.J. Structure, function and evolution of the hemagglutinin-esterase proteins of corona- and toroviruses. Glycoconj. J. 2006, 23, 59–72. [Google Scholar] [CrossRef]
- Desforges, M.; Desjardins, J.; Zhang, C.; Talbot, P.J. The acetyl-esterase activity of the hemagglutinin-esterase protein of human coronavirus OC43 strongly enhances the production of infectious virus. J. Virol. 2013, 87, 3097–3107. [Google Scholar] [CrossRef] [Green Version]
- Langereis, M.A.; Bakkers, M.J.; Deng, L.; Padler-Karavani, V.; Vervoort, S.J.; Hulswit, R.J.; van Vliet, A.L.; Gerwig, G.J.; de Poot, S.A.; Boot, W.; et al. Complexity and Diversity of the Mammalian Sialome Revealed by Nidovirus Virolectins. Cell Rep. 2015, 11, 1966–1978. [Google Scholar] [CrossRef] [Green Version]
- Bakkers, M.J.; Lang, Y.; Feitsma, L.J.; Hulswit, R.J.; de Poot, S.A.; van Vliet, A.L.; Margine, I.; de Groot-Mijnes, J.D.; van Kuppeveld, F.J.; Langereis, M.A.; et al. Betacoronavirus Adaptation to Humans Involved Progressive Loss of Hemagglutinin-Esterase Lectin Activity. Cell Host Microbe 2017, 21, 356–366. [Google Scholar] [CrossRef] [Green Version]
- Lang, Y.; Li, W.; Li, Z.; Koerhuis, D.; van den Burg, A.C.S.; Rozemuller, E.; Bosch, B.J.; van Kuppeveld, F.J.M.; Boons, G.J.; Huizinga, E.G.; et al. Coronavirus hemagglutinin-esterase and spike proteins coevolve for functional balance and optimal virion avidity. Proc. Natl. Acad. Sci. USA 2020, 117, 25759–25770. [Google Scholar] [CrossRef]
- Kunkel, F.; Herrler, G. Structural and functional analysis of the surface protein of human coronavirus OC43. Virology 1993, 195, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Stodola, J.K.; Dubois, G.; Le Coupanec, A.; Desforges, M.; Talbot, P.J. The OC43 human coronavirus envelope protein is critical for infectious virus production and propagation in neuronal cells and is a determinant of neurovirulence and CNS pathology. Virology 2018, 515, 134–149. [Google Scholar] [CrossRef] [PubMed]
- Mounir, S.; Talbot, P.J. Molecular characterization of the S protein gene of human coronavirus OC43. J. Gen. Virol. 1993, 74 Pt 9, 1981–1987. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Zhang, Y.; Li, J.; Xiao, Y.; Zhang, J.; Wang, Y.; Chen, L.; Paranhos-Baccala, G.; Wang, J. Genetic drift of human coronavirus OC43 spike gene during adaptive evolution. Sci. Rep. 2015, 5, 11451. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Wang, K.; Ping, X.; Yu, W.; Qian, Z.; Xiong, S.; Sun, B. The ns12.9 Accessory Protein of Human Coronavirus OC43 Is a Viroporin Involved in Virion Morphogenesis and Pathogenesis. J. Virol. 2015, 89, 11383–11395. [Google Scholar] [CrossRef] [Green Version]
- Kamahora, T.; Soe, L.H.; Lai, M.M. Sequence analysis of nucleocapsid gene and leader RNA of human coronavirus OC43. Virus Res. 1989, 12, 1–9. [Google Scholar] [CrossRef]
- Sturman, L.S.; Holmes, K.V.; Behnke, J. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J. Virol. 1980, 33, 449–462. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.Y.; Hsu, Y.L.; Chiang, W.L.; Hou, M.H. Elucidation of the stability and functional regions of the human coronavirus OC43 nucleocapsid protein. Protein Sci. 2009, 18, 2209–2218. [Google Scholar] [CrossRef] [Green Version]
- Chen, I.J.; Chou, C.C.; Liu, C.L.; Lee, C.C.; Kan, L.S.; Hou, M.H. Crystallization and preliminary X-ray diffraction analysis of the N-terminal domain of human coronavirus OC43 nucleocapsid protein. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 815–818. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, C.M.; Gebauer, F.; Sune, C.; Mendez, A.; Dopazo, J.; Enjuanes, L. Genetic evolution and tropism of transmissible gastroenteritis coronaviruses. Virology 1992, 190, 92–105. [Google Scholar] [CrossRef]
- Lam, T.T.; Jia, N.; Zhang, Y.W.; Shum, M.H.; Jiang, J.F.; Zhu, H.C.; Tong, Y.G.; Shi, Y.X.; Ni, X.B.; Liao, Y.S.; et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 2020, 583, 282–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vijgen, L.; Keyaerts, E.; Lemey, P.; Maes, P.; Van Reeth, K.; Nauwynck, H.; Pensaert, M.; Van Ranst, M. Evolutionary history of the closely related group 2 coronaviruses: Porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J. Virol. 2006, 80, 7270–7274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, S.K.; Li, K.S.; Huang, Y.; Shek, C.T.; Tse, H.; Wang, M.; Choi, G.K.; Xu, H.; Lam, C.S.; Guo, R.; et al. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J. Virol. 2010, 84, 2808–2819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hogue, B.G.; King, B.; Brian, D.A. Antigenic relationships among proteins of bovine coronavirus, human respiratory coronavirus OC43, and mouse hepatitis coronavirus A59. J. Virol. 1984, 51, 384–388. [Google Scholar] [CrossRef] [Green Version]
- Lapps, W.; Brian, D.A. Oligonucleotide fingerprints of antigenically related bovine coronavirus and human coronavirus OC43. Arch. Virol. 1985, 86, 101–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maurier, F.; Beury, D.; Flechon, L.; Varre, J.S.; Touzet, H.; Goffard, A.; Hot, D.; Caboche, S. A complete protocol for whole-genome sequencing of virus from clinical samples: Application to coronavirus OC43. Virology 2019, 531, 141–148. [Google Scholar] [CrossRef]
- Vieler, E.; Schlapp, T.; Anders, C.; Herbst, W. Genomic relationship of porcine hemagglutinating encephalomyelitis virus to bovine coronavirus and human coronavirus OC43 as studied by the use of bovine coronavirus S gene-specific probes. Arch. Virol. 1995, 140, 1215–1223. [Google Scholar] [CrossRef] [Green Version]
- Mounir, S.; Talbot, P.J. Human coronavirus OC43 RNA 4 lacks two open reading frames located downstream of the S gene of bovine coronavirus. Virology 1993, 192, 355–360. [Google Scholar] [CrossRef]
- Kin, N.; Miszczak, F.; Lin, W.; Gouilh, M.A.; Vabret, A.; Consortium, E. Genomic Analysis of 15 Human Coronaviruses OC43 (HCoV-OC43s) Circulating in France from 2001 to 2013 Reveals a High Intra-Specific Diversity with New Recombinant Genotypes. Viruses 2015, 7, 2358–2377. [Google Scholar] [CrossRef] [Green Version]
- Kistler, K.E.; Bedford, T. Evidence for adaptive evolution in the receptor-binding domain of seasonal coronaviruses OC43 and 229E. Elife 2021, 10, e64509. [Google Scholar] [CrossRef]
- Krempl, C.; Schultze, B.; Herrler, G. Analysis of cellular receptors for human coronavirus OC43. Adv. Exp. Med. Biol. 1995, 380, 371–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlasak, R.; Luytjes, W.; Spaan, W.; Palese, P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc. Natl. Acad. Sci. USA 1988, 85, 4526–4529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Dong, W.; Milewska, A.; Golda, A.; Qi, Y.; Zhu, Q.K.; Marasco, W.A.; Baric, R.S.; Sims, A.C.; Pyrc, K.; et al. Human Coronavirus HKU1 Spike Protein Uses O-Acetylated Sialic Acid as an Attachment Receptor Determinant and Employs Hemagglutinin-Esterase Protein as a Receptor-Destroying Enzyme. J. Virol. 2015, 89, 7202–7213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tortorici, M.A.; Walls, A.C.; Lang, Y.; Wang, C.; Li, Z.; Koerhuis, D.; Boons, G.J.; Bosch, B.J.; Rey, F.A.; de Groot, R.J.; et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 2019, 26, 481–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, A.R. HLA class I antigen serves as a receptor for human coronavirus OC43. Immunol. Investig. 1993, 22, 95–103. [Google Scholar] [CrossRef] [Green Version]
- Owczarek, K.; Szczepanski, A.; Milewska, A.; Baster, Z.; Rajfur, Z.; Sarna, M.; Pyrc, K. Early events during human coronavirus OC43 entry to the cell. Sci. Rep. 2018, 8, 7124. [Google Scholar] [CrossRef] [Green Version]
- Davies, J.P.; Almasy, K.M.; McDonald, E.F.; Plate, L. Comparative Multiplexed Interactomics of SARS-CoV-2 and Homologous Coronavirus Nonstructural Proteins Identifies Unique and Shared Host-Cell Dependencies. ACS Infect. Dis. 2020, 6, 3174–3189. [Google Scholar] [CrossRef]
- Hoffmann, H.H.; Sanchez-Rivera, F.J.; Schneider, W.M.; Luna, J.M.; Soto-Feliciano, Y.M.; Ashbrook, A.W.; Le Pen, J.; Leal, A.A.; Ricardo-Lax, I.; Michailidis, E.; et al. Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors. Cell Host Microbe 2020, 29, 267–280.e5. [Google Scholar] [CrossRef]
- Wang, R.; Simoneau, C.R.; Kulsuptrakul, J.; Bouhaddou, M.; Travisano, K.A.; Hayashi, J.M.; Carlson-Stevermer, J.; Zengel, J.R.; Richards, C.M.; Fozouni, P.; et al. Genetic Screens Identify Host Factors for SARS-CoV-2 and Common Cold Coronaviruses. Cell 2021, 184, 106–119.e114. [Google Scholar] [CrossRef]
- Schneider, W.M.; Luna, J.M.; Hoffmann, H.H.; Sanchez-Rivera, F.J.; Leal, A.A.; Ashbrook, A.W.; Le Pen, J.; Ricardo-Lax, I.; Michailidis, E.; Peace, A.; et al. Genome-Scale Identification of SARS-CoV-2 and Pan-coronavirus Host Factor Networks. Cell 2021, 184, 120–132.e114. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef] [Green Version]
- Collins, A.R. Interferon gamma potentiates human coronavirus OC43 infection of neuronal cells by modulation of HLA class I expression. Immunol. Investig. 1995, 24, 977–986. [Google Scholar] [CrossRef] [PubMed]
- Lai, F.W.; Stephenson, K.B.; Mahony, J.; Lichty, B.D. Human coronavirus OC43 nucleocapsid protein binds microRNA 9 and potentiates NF-kappaB activation. J. Virol. 2014, 88, 54–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Guo, F.; Liu, F.; Cuconati, A.; Chang, J.; Block, T.M.; Guo, J.T. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc. Natl. Acad. Sci. USA 2014, 111, 6756–6761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loo, S.L.; Wark, P.A.B.; Esneau, C.; Nichol, K.S.; Hsu, A.C.; Bartlett, N.W. Human coronaviruses 229E and OC43 replicate and induce distinct antiviral responses in differentiated primary human bronchial epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2020, 319, L926–L931. [Google Scholar] [CrossRef] [PubMed]
- Hicks, J.; Klumpp-Thomas, C.; Kalish, H.; Shunmugavel, A.; Mehalko, J.; Denson, J.P.; Snead, K.; Drew, M.; Corbett, K.; Graham, B.; et al. Serologic cross-reactivity of SARS-CoV-2 with endemic and seasonal Betacoronaviruses. medRxiv 2020. [Google Scholar] [CrossRef]
- Dugas, M.; Grote-Westrick, T.; Vollenberg, R.; Lorentzen, E.; Brix, T.; Schmidt, H.; Tepasse, P.R.; Kuhn, J. Less severe course of COVID-19 is associated with elevated levels of antibodies against seasonal human coronaviruses OC43 and HKU1 (HCoV OC43, HCoV HKU1). Int. J. Infect. Dis. 2021, 105, 304–306. [Google Scholar] [CrossRef]
- Saletti, G.; Gerlach, T.; Jansen, J.M.; Molle, A.; Elbahesh, H.; Ludlow, M.; Li, W.; Bosch, B.J.; Osterhaus, A.; Rimmelzwaan, G.F. Older adults lack SARS CoV-2 cross-reactive T lymphocytes directed to human coronaviruses OC43 and NL63. Sci. Rep. 2020, 10, 21447. [Google Scholar] [CrossRef]
- Lee, C.H.; Pinho, M.P.; Buckley, P.R.; Woodhouse, I.B.; Ogg, G.; Simmons, A.; Napolitani, G.; Koohy, H. Potential CD8+ T Cell Cross-Reactivity Against SARS-CoV-2 Conferred by Other Coronavirus Strains. Front. Immunol. 2020, 11, 579480. [Google Scholar] [CrossRef]
- Patrick, D.M.; Petric, M.; Skowronski, D.M.; Guasparini, R.; Booth, T.F.; Krajden, M.; McGeer, P.; Bastien, N.; Gustafson, L.; Dubord, J.; et al. An Outbreak of Human Coronavirus OC43 Infection and Serological Cross-reactivity with SARS Coronavirus. Can. J. Infect. Dis. Med. Microbiol. 2006, 17, 330–336. [Google Scholar] [CrossRef]
- Braun, J.; Loyal, L.; Frentsch, M.; Wendisch, D.; Georg, P.; Kurth, F.; Hippenstiel, S.; Dingeldey, M.; Kruse, B.; Fauchere, F.; et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 2020, 587, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Mateus, J.; Grifoni, A.; Tarke, A.; Sidney, J.; Ramirez, S.I.; Dan, J.M.; Burger, Z.C.; Rawlings, S.A.; Smith, D.M.; Phillips, E.; et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science 2020, 370, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e1415. [Google Scholar] [CrossRef] [PubMed]
- Arbour, N.; Ekande, S.; Cote, G.; Lachance, C.; Chagnon, F.; Tardieu, M.; Cashman, N.R.; Talbot, P.J. Persistent infection of human oligodendrocytic and neuroglial cell lines by human coronavirus 229E. J. Virol. 1999, 73, 3326–3337.e1415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arbour, N.; Cote, G.; Lachance, C.; Tardieu, M.; Cashman, N.R.; Talbot, P.J. Acute and persistent infection of human neural cell lines by human coronavirus OC43. J. Virol. 1999, 73, 3338–3350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, A.R.; Sorensen, O. Regulation of viral persistence in human glioblastoma and rhabdomyosarcoma cells infected with coronavirus OC43. Microb. Pathog. 1986, 1, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Bonavia, A.; Arbour, N.; Yong, V.W.; Talbot, P.J. Infection of primary cultuRes. of human neural cells by human coronaviruses 229E and OC43. J. Virol. 1997, 71, 800–806. [Google Scholar] [CrossRef] [Green Version]
- Pearson, J.; Mims, C.A. Differential susceptibility of cultured neural cells to the human coronavirus OC43. J. Virol. 1985, 53, 1016–1019. [Google Scholar] [CrossRef] [Green Version]
- Edwards, J.A.; Denis, F.; Talbot, P.J. Activation of glial cells by human coronavirus OC43 infection. J. Neuroimmunol. 2000, 108, 73–81. [Google Scholar] [CrossRef] [Green Version]
- Jacomy, H.; Talbot, P.J. HCoV-OC43-induced apoptosis of murine neuronal cells. Adv. Exp. Med. Biol. 2006, 581, 473–478. [Google Scholar] [CrossRef]
- Desforges, M.; Le Coupanec, A.; Dubeau, P.; Bourgouin, A.; Lajoie, L.; Dube, M.; Talbot, P.J. Human Coronaviruses and Other Respiratory Viruses: Underestimated Opportunistic Pathogens of the Central Nervous System? Viruses 2019, 12, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacomy, H.; St-Jean, J.R.; Brison, E.; Marceau, G.; Desforges, M.; Talbot, P.J. Mutations in the spike glycoprotein of human coronavirus OC43 modulate disease in BALB/c mice from encephalitis to flaccid paralysis and demyelination. J. Neurovirol. 2010, 16, 279–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meessen-Pinard, M.; Le Coupanec, A.; Desforges, M.; Talbot, P.J. Pivotal Role of Receptor-Interacting Protein Kinase 1 and Mixed Lineage Kinase Domain-Like in Neuronal Cell Death Induced by the Human Neuroinvasive Coronavirus OC43. J. Virol. 2017, 91, e01513-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearson, J.; Mims, C.A. Selective vulnerability of neural cells and age-related susceptibility to OC43 virus in mice. Arch. Virol. 1983, 77, 109–118. [Google Scholar] [CrossRef]
- Jacomy, H.; Talbot, P.J. Susceptibility of murine CNS to OC43 infection. Adv. Exp. Med. Biol. 2001, 494, 101–107. [Google Scholar] [CrossRef]
- Dube, M.; Le Coupanec, A.; Wong, A.H.M.; Rini, J.M.; Desforges, M.; Talbot, P.J. Axonal Transport Enables Neuron-to-Neuron Propagation of Human Coronavirus OC43. J. Virol. 2018, 92, e00404-18. [Google Scholar] [CrossRef] [Green Version]
- Butler, N.; Pewe, L.; Trandem, K.; Perlman, S. HCoV-OC43-induced encephalitis is in part immune-mediated. Adv. Exp. Med. Biol. 2006, 581, 531–534. [Google Scholar]
- Favreau, D.J.; Desforges, M.; St-Jean, J.R.; Talbot, P.J. A human coronavirus OC43 variant harboring persistence-associated mutations in the S glycoprotein differentially induces the unfolded protein response in human neurons as compared to wild-type virus. Virology 2009, 395, 255–267. [Google Scholar] [CrossRef]
- Niu, J.; Shen, L.; Huang, B.; Ye, F.; Zhao, L.; Wang, H.; Deng, Y.; Tan, W. Non-invasive bioluminescence imaging of HCoV-OC43 infection and therapy in the central nervous system of live mice. Antivir. Res. 2020, 173, 104646. [Google Scholar] [CrossRef]
- Le Coupanec, A.; Desforges, M.; Kaufer, B.; Dubeau, P.; Cote, M.; Talbot, P.J. Potential differences in cleavage of the S protein and type-1 interferon together control human coronavirus infection, propagation, and neuropathology within the central nervous system. J. Virol. 2021, 95, e00140-21. [Google Scholar] [CrossRef]
- Yeh, E.A.; Collins, A.; Cohen, M.E.; Duffner, P.K.; Faden, H. Detection of coronavirus in the central nervous system of a child with acute disseminated encephalomyelitis. Pediatrics 2004, 113, e73–e76. [Google Scholar] [CrossRef] [Green Version]
- Morfopoulou, S.; Brown, J.R.; Davies, E.G.; Anderson, G.; Virasami, A.; Qasim, W.; Chong, W.K.; Hubank, M.; Plagnol, V.; Desforges, M.; et al. Human Coronavirus OC43 Associated with Fatal Encephalitis. N. Engl. J. Med. 2016, 375, 497–498. [Google Scholar] [CrossRef]
- Kasereka, M.C.; Hawkes, M.T. Neuroinvasive potential of human coronavirus OC43: Case report of fatal encephalitis in an immunocompromised host. J. Neurovirol. 2021, 27, 340–344. [Google Scholar] [CrossRef]
- Arbour, N.; Day, R.; Newcombe, J.; Talbot, P.J. Neuroinvasion by human respiratory coronaviruses. J. Virol. 2000, 74, 8913–8921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hovanec, D.L.; Flanagan, T.D. Detection of antibodies to human coronaviruses 229E and OC43 in the sera of multiple sclerosis patients and normal subjects. Infect. Immun. 1983, 41, 426–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debiaggi, M.; Perduca, M.; Romero, E.; Cereda, P.M. Phosphatidyl-serine inhibition of OC43 and NCDCV coronavirus infectivity. Microbiologica 1985, 8, 313–317. [Google Scholar] [PubMed]
- Collins, A.R.; Grubb, A. Inhibitory effects of recombinant human cystatin C on human coronaviruses. Antimicrob. Agents Chemother. 1991, 35, 2444–2446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, A.R.; Grubb, A. Cystatin D, a natural salivary cysteine protease inhibitor, inhibits coronavirus replication at its physiologic concentration. Oral Microbiol. Immunol. 1998, 13, 59–61. [Google Scholar] [CrossRef]
- Shen, L.; Yang, Y.; Ye, F.; Liu, G.; Desforges, M.; Talbot, P.J.; Tan, W. Safe and Sensitive Antiviral Screening Platform Based on Recombinant Human Coronavirus OC43 Expressing the Luciferase Reporter Gene. Antimicrob. Agents Chemother. 2016, 60, 5492–5503. [Google Scholar] [CrossRef] [Green Version]
- Keyaerts, E.; Li, S.; Vijgen, L.; Rysman, E.; Verbeeck, J.; Van Ranst, M.; Maes, P. Antiviral activity of chloroquine against human coronavirus OC43 infection in newborn mice. Antimicrob. Agents Chemother. 2009, 53, 3416–3421. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, S.; Wang, K.; Yu, W.; Sun, B.; Schwarz, W. Emodin inhibits current through SARS-associated coronavirus 3a protein. Antivir. Res. 2011, 90, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Brison, E.; Jacomy, H.; Desforges, M.; Talbot, P.J. Novel Treatment with Neuroprotective and Antiviral Properties against a Neuroinvasive Human Respiratory Virus. J. Virol. 2014, 88, 1548–1563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milewska, A.; Kaminski, K.; Ciejka, J.; Kosowicz, K.; Zeglen, S.; Wojarski, J.; Nowakowska, M.; Szczubialka, K.; Pyrc, K. HTCC: Broad Range Inhibitor of Coronavirus Entry. PLoS ONE 2016, 11, e0156552. [Google Scholar] [CrossRef] [Green Version]
- Milewska, A.; Chi, Y.; Szczepanski, A.; Barreto-Duran, E.; Dabrowska, A.; Botwina, P.; Obloza, M.; Liu, K.; Liu, D.; Guo, X.; et al. HTCC as a Polymeric Inhibitor of SARS-CoV-2 and MERS-CoV. J. Virol. 2021, 95, e01622-20. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Niu, J.; Wang, C.; Huang, B.; Wang, W.; Zhu, N.; Deng, Y.; Wang, H.; Ye, F.; Cen, S.; et al. High-Throughput Screening and Identification of Potent Broad-Spectrum Inhibitors of Coronaviruses. J. Virol. 2019, 93, e00023-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.E.; Min, J.S.; Jang, M.S.; Lee, J.Y.; Shin, Y.S.; Song, J.H.; Kim, H.R.; Kim, S.; Jin, Y.H.; Kwon, S. Natural Bis-Benzylisoquinoline Alkaloids-Tetrandrine, Fangchinoline, and Cepharanthine, Inhibit Human Coronavirus OC43 Infection of MRC-5 Human Lung Cells. Biomolecules 2019, 9, 696. [Google Scholar] [CrossRef] [Green Version]
- Zafferani, M.; Haddad, C.; Luo, L.; Davila-Calderon, J.; Yuan-Chiu, L.; Shema Mugisha, C.; Monaghan, A.G.; Kennedy, A.A.; Yesselman, J.D.; Gifford, R.R.; et al. Amilorides inhibit SARS-CoV-2 replication in vitro by targeting RNA structures. bioRxiv 2020. [Google Scholar] [CrossRef]
- Yang, C.W.; Lee, Y.Z.; Hsu, H.Y.; Jan, J.T.; Lin, Y.L.; Chang, S.Y.; Peng, T.T.; Yang, R.B.; Liang, J.J.; Liao, C.C.; et al. Inhibition of SARS-CoV-2 by Highly Potent Broad-Spectrum Anti-Coronaviral Tylophorine-Based Derivatives. Front. Pharm. 2020, 11, 606097. [Google Scholar] [CrossRef]
- Min, J.S.; Kim, D.E.; Jin, Y.H.; Kwon, S. Kurarinone Inhibits HCoV-OC43 Infection by Impairing the Virus-Induced Autophagic Flux in MRC-5 Human Lung Cells. J. Clin. Med. 2020, 9, 2230. [Google Scholar] [CrossRef]
- Bleasel, M.D.; Peterson, G.M. Emetine, Ipecac, Ipecac Alkaloids and Analogues as Potential Antiviral Agents for Coronaviruses. Pharmaceuticals 2020, 13, 51. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.W.; Peng, T.T.; Hsu, H.Y.; Lee, Y.Z.; Wu, S.H.; Lin, W.H.; Ke, Y.Y.; Hsu, T.A.; Yeh, T.K.; Huang, W.Z.; et al. Repurposing old drugs as antiviral agents for coronaviruses. Biomed. J. 2020, 43, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Marcello, A.; Civra, A.; Milan Bonotto, R.; Nascimento Alves, L.; Rajasekharan, S.; Giacobone, C.; Caccia, C.; Cavalli, R.; Adami, M.; Brambilla, P.; et al. The cholesterol metabolite 27-hydroxycholesterol inhibits SARS-CoV-2 and is markedly decreased in COVID-19 patients. Redox Biol. 2020, 36, 101682. [Google Scholar] [CrossRef] [PubMed]
- Good, S.S.; Westover, J.; Jung, K.H.; Zhou, X.J.; Moussa, A.; La Colla, P.; Collu, G.; Canard, B.; Sommadossi, J.P. AT-527, a double prodrug of a guanosine nucleotide analog, is a potent inhibitor of SARS-CoV-2 in vitro and a promising oral antiviral for treatment of COVID-19. Antimicrob. Agents Chemother. 2021, 65, e02479-20. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.; Park, R.; Park, Y.I.; Cha, Y.E.; Yamamoto, A.; Lee, J.I.; Park, J. EGCG, a green tea polyphenol, inhibits human coronavirus replication in vitro. Biochem. Biophys. Res. Commun. 2021, 547, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Meng, X.; Zhang, F.; Xiang, Y.; Wang, J. The in vitro antiviral activity of lactoferrin against common human coronaviruses and SARS-CoV-2 is mediated by targeting the heparan sulfate co-receptor. Emerg. Microbes Infect. 2021, 10, 317–330. [Google Scholar] [CrossRef] [PubMed]

| Structural Protein | Key Function | References |
|---|---|---|
| Hemagglutinin-esterase | Receptor-binding and degradation | [52,53,54,55,56] |
| Spike protein | Receptor-binding and hemagglutination | [61] |
| Envelope protein | Ion channel formation | [62] |
| Membrane protein | Virion morphogenesis | [45] |
| Nucleocapsid | Helical nucleocapsid formation | [1] |
| Host Factor or Process | Steps of Virus Life Cycle | References |
|---|---|---|
| 9-O-acetyl sialic acid | Host receptor binding | [6,82,83,84] |
| MHC class I | Host receptor binding | [18,85] |
| Caveolin-1-dependent endocytosis | Virus entry | [86] |
| ER structure | Virus RNA replication | [87] |
| Rab GTPase and glycosylphosphatidylinositol | Virus assembly and trafficking | [88] |
| Endosome maturation, phosphatidylinositol phosphate, and cholesterol homeostasis | Virus assembly and trafficking | [89] |
| VMP1, TMEM41B, and TMEM64 | ER membrane remodeling | [90] |
| Dynamin-dependent budding | Virus exit | [86] |
| Name | Experiment Type | EC50 (mM) * | References |
|---|---|---|---|
| Phosphatidyl-serine | in vitro | na | [126] |
| Cystatin C and D | in vitro | 0.8 | [127,128] |
| Chloroquine | in vitro and in vivo | 0.33 | [129] |
| Emodin | in vitro (inhibition of 3a ion channel) | <10 | [131] |
| Memantine | in vivo | NA ** | [132] |
| HTCC | in vitro | NA | [133] |
| Lycorine | in vivo | <5 | [135] |
| Bis-benzylisoquinoline alkaloids | in vitro | <0.1 | [136] |
| Amiloride | in vitro | >10 | [137] |
| Tylophorine-based compounds | in vitro | 0.1–1 | [138] |
| Cardenolides and bufadienolides | in vitro | 0.1–1 | [138] |
| Kurarinone | in vitro | 3.458 | [139] |
| Emetine | in vitro | 0.21 | [140,141] |
| Oxysterol 27-hydroxycholesterol | in vitro | <10 | [142] |
| Valinomycin | in vitro | 6.15 | [40] |
| AT-527 | in vitro | 2.2 | [143] |
| EGCG | in vitro | 14.6 | [144] |
| Lactoferrin | in vitro | <50 mg/mL | [145] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.I.; Lee, C. Human Coronavirus OC43 as a Low-Risk Model to Study COVID-19. Viruses 2023, 15, 578. https://doi.org/10.3390/v15020578
Kim MI, Lee C. Human Coronavirus OC43 as a Low-Risk Model to Study COVID-19. Viruses. 2023; 15(2):578. https://doi.org/10.3390/v15020578
Chicago/Turabian StyleKim, Mi Il, and Choongho Lee. 2023. "Human Coronavirus OC43 as a Low-Risk Model to Study COVID-19" Viruses 15, no. 2: 578. https://doi.org/10.3390/v15020578
APA StyleKim, M. I., & Lee, C. (2023). Human Coronavirus OC43 as a Low-Risk Model to Study COVID-19. Viruses, 15(2), 578. https://doi.org/10.3390/v15020578






