The Effect of a Bacillus Probiotic and Essential Oils Compared to an Ionophore on the Rumen Microbiome Composition of Feedlot Cattle
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Performance Results and VFA Concentrations
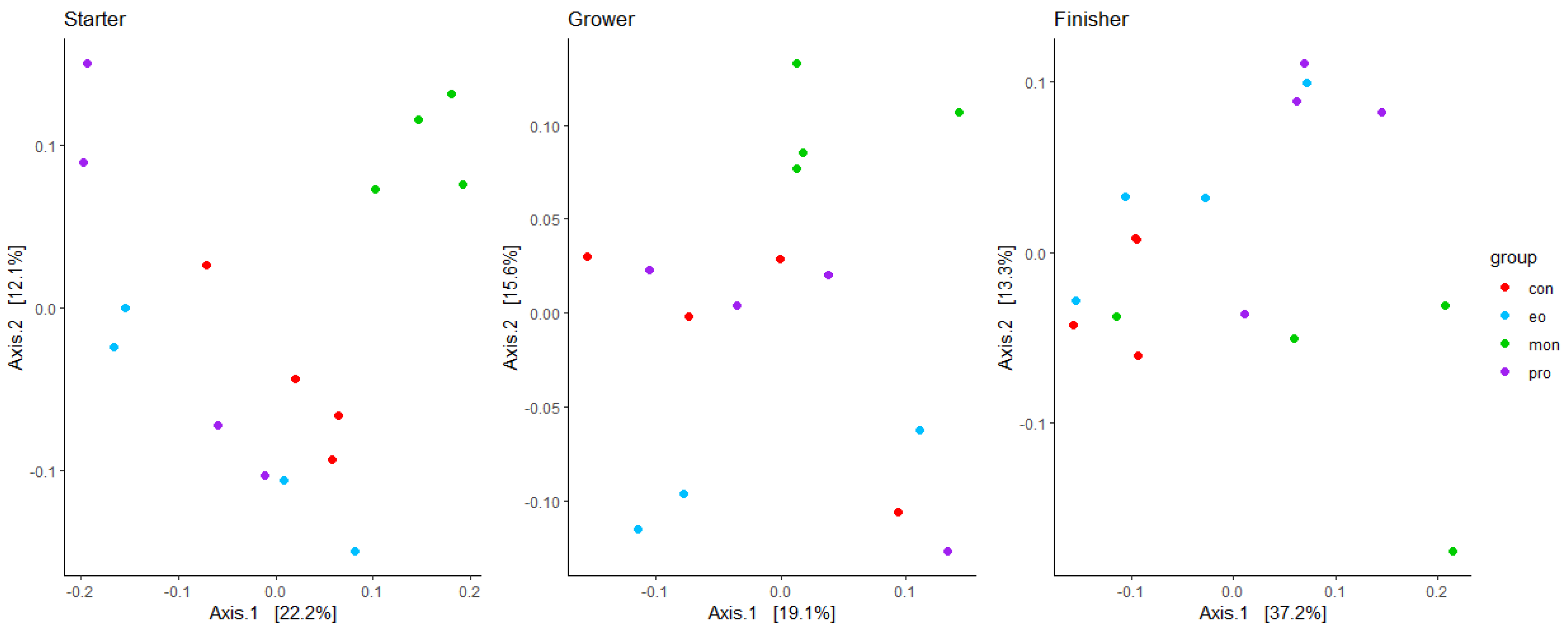
3.2. Alpha and Beta Diversity of the Rumen Microbial Composition
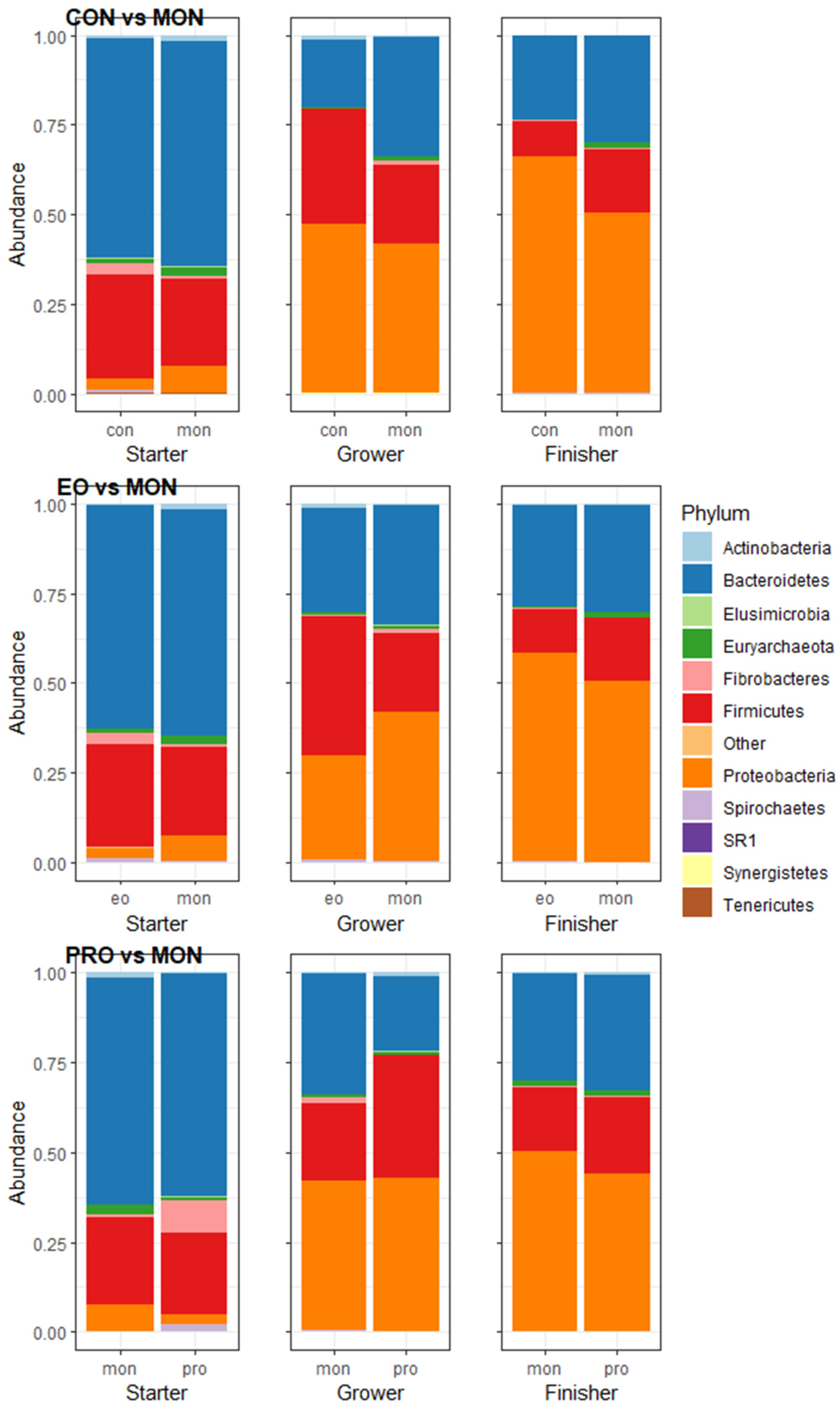
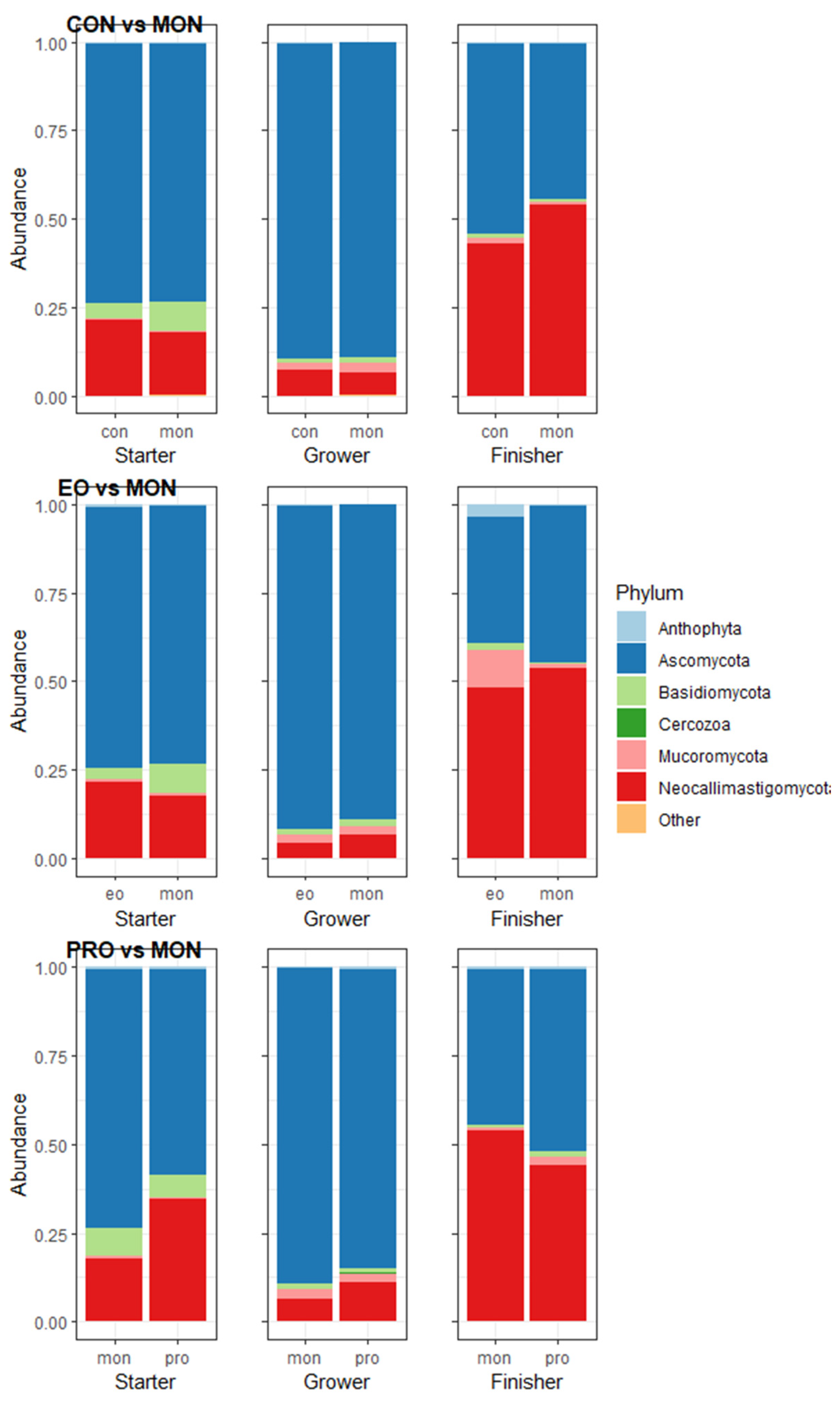
3.3. Rumen Microorganism Abundances
4. Discussion
4.1. Monensin vs. Control
4.2. Monensin vs. Essential Oils
4.3. Monensin vs. Probiotic
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Myer, P.R.; Smith, T.P.L.L.; Wells, J.E.; Kuehn, L.A.; Freetly, H.C. Rumen Microbiome from Steers Differing in Feed Efficiency. PLoS ONE 2015, 10, e0129174. [Google Scholar] [CrossRef] [PubMed]
- Freetly, H.C.; Dickey, A.; Lindholm-Perry, A.K.; Richard, M.; Keele, J.W.; Foote, A.P.; Wells, J.E. Digestive Tract Microbiota of Beef Cattle That Differed in Feed Efficiency. J. Anim. Sci. 2020, 98, skaa008. [Google Scholar] [CrossRef] [PubMed]
- Perea, K.; Perz, K.; Olivo, S.K.; Williams, A.; Lachman, M.; Ishaq, S.L.; Thomson, J.; Yeoman, C.J. Feed Efficiency Phenotypes in Lambs Involve Changes in Ruminal, Colonic, and Small-Intestine-Located Microbiota. J. Anim. Sci. 2017, 95, 2585–2592. [Google Scholar] [CrossRef] [PubMed]
- Jouany, J.; Morgavi, D.P. Use of ‘Natural’ Products as Alternatives to Antibiotic Feed Additives in Ruminant Production. Animal 2007, 1, 1443–1466. [Google Scholar] [CrossRef] [PubMed]
- Duffield, T.F.; Merrill, J.K.; Bagg, R.N. Meta-Analysis of the Effects of Monensin in Beef Cattle on Feed Efficiency, Body Weight Gain, and Dry Matter Intake. J. Anim. Sci. 2012, 90, 4583–4592. [Google Scholar] [CrossRef] [PubMed]
- Markowiak, P.; Śliżewska, K. The Role of Probiotics, Prebiotics and Synbiotics in Animal Nutrition. Gut Pathog. 2018, 10, 21. [Google Scholar] [CrossRef] [PubMed]
- Du, R.; Jiao, S.; Dai, Y.; An, J.; Lv, J.; Yan, X.; Wang, J.; Han, B. Probiotic Bacillus Amyloliquefaciens C-1 Improves Growth Performance, Stimulates GH/IGF-1, and Regulates the Gut Microbiota of Growth-Retarded Beef Calves. Front. Microbiol. 2018, 9, 2006. [Google Scholar] [CrossRef]
- Sun, P.; Wang, J.Q.; Deng, L.F. Effects of Bacillus Subtilis Natto on Milk Production, Rumen Fermentation and Ruminal Microbiome of Dairy Cows. Animal 2013, 7, 216–222. [Google Scholar] [CrossRef]
- Hosoi, T.; Ametani, A.; Kiuchi, K.; Kaminogawa, S. Improved Growth and Viability of Lactobacilli in the Presence of Bacillus Subtilis (Natto), Catalase, or Subtilisin. Can. J. Microbiol. 2000, 46, 892–897. [Google Scholar] [CrossRef]
- Khochamit, N.; Siripornadulsil, S.; Sukon, P.; Siripornadulsil, W. Antibacterial Activity and Genotypic-Phenotypic Characteristics of Bacteriocin-Producing Bacillus Subtilis KKU213: Potential as a Probiotic Strain. Microbiol. Res. 2015, 170, 36–50. [Google Scholar] [CrossRef]
- Patra, A.K.; Yu, Z. Effects of Essential Oils on Methane Production and Fermentation by, and Abundance and Diversity of, Rumen Microbial Populations. Appl. Environ. Microbiol. 2012, 78, 4271–4280. [Google Scholar] [CrossRef] [PubMed]
- Calsamiglia, S.; Busquet, M.; Cardozo, P.W.; Castillejos, L.; Ferret, A. Invited Review: Essential Oils as Modifiers of Rumen Microbial Fermentation. J. Dairy Sci. 2007, 90, 2580–2595. [Google Scholar] [CrossRef] [PubMed]
- Cardozo, P.W.; Calsamiglia, S.; Ferret, A.; Kamel, C. Effects of Alfalfa Extract, Anise, Capsicum, and a Mixture of Cinnamaldehyde and Eugenol on Ruminal Fermentation and Protein Degradation in Beef Heifers Fed a High-Concentrate Diet. J. Anim. Sci. 2006, 84, 2801–2808. [Google Scholar] [CrossRef] [PubMed]
- Benchaar, C.; Duynisveld, J.L.; Charmley, E. Effects of Monensin and Increasing Dose Levels of a Mixture of Essential Oil Compounds on Intake, Digestion and Growth Performance of Beef Cattle. Can. J. Anim. Sci. 2006, 86, 91–96. [Google Scholar]
- Meschiatti, M.A.P.; Gouvêa, V.N.; Pellarin, L.A.; Batalha, C.D.A.; Biehl, M.V.; Acedo, T.S.; Dórea, J.R.R.; Tamassia, L.F.M.; Owens, F.N.; Santos, F.A.P. Feeding the Combination of Essential Oils and Exogenous α-Amylase Increases Performance and Carcass Production of Finishing Beef Cattle. J. Anim. Sci. 2019, 97, 456–471. [Google Scholar] [CrossRef]
- Schären, M.; Drong, C.; Kiri, K.; Riede, S.; Gardener, M.; Meyer, U.; Hummel, J.; Urich, T.; Breves, G.; Dänicke, S. Differential Effects of Monensin and a Blend of Essential Oils on Rumen Microbiota Composition of Transition Dairy Cows. J. Dairy Sci. 2016, 100, 2765–2783. [Google Scholar] [CrossRef]
- Linde, D.A.; Toit, C.J.L.; Scholtz, M.M.; Schokker, D. Rumen Microbial Diversity of Bonsmara Cattle Using Amplicon Sequencing during a 120-Day Growth Trial. S. Afr. J. Anim. Sci. 2022, 52, 148–161. [Google Scholar]
- Wu, X.; Huang, S.; Huang, J.; Peng, P.; Liu, Y.; Han, B.; Sun, D. Identification of the Potential Role of the Rumen Microbiome in Milk Protein and Fat Synthesis in Dairy Cows Using Metagenomic Sequencing. Animals 2021, 11, 1247. [Google Scholar] [CrossRef]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L.; Author, N.B. Differential Analysis of Gene Regulation at Transcript Resolution with RNA-Seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef]
- Paz, H.A.; Anderson, C.L.; Muller, M.J.; Kononoff, P.J.; Fernando, S.C. Rumen Bacterial Community Composition in Holstein and Jersey Cows Is Different under Same Dietary Condition and Is Not Affected by Sampling Method. Front. Microbiol. 2016, 7, 1206. [Google Scholar] [CrossRef]
- Prates, A.; de Oliveira, J.A.; Abecia, L.; Fondevila, M. Effects of Preservation Procedures of Rumen Inoculum on in Vitro Microbial Diversity and Fermentation. Anim. Feed Sci. Technol. 2010, 155, 186–193. [Google Scholar] [CrossRef]
- Balthrop, J.; Brand, B.; Cowie, R.A.; Danier, J.; De Boever, J.; de Jonge, L.; Jackson, F.; Makkar, H.P.S.; Piotrowski, C. FAO Quality Assurance for Animal Feed Analysis Laboratories; Food and Agriculture Organization of the United Nations: Rome, Italy, 2011; ISBN 9789251070505. [Google Scholar]
- Terré, M.; Castells, L.; Fàbregas, F.; Bach, A. Short Communication: Comparison of PH, Volatile Fatty Acids, and Microbiome of Rumen Samples from Preweaned Calves Obtained via Cannula or Stomach Tube. J. Dairy Sci. 2013, 96, 5290–5294. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and Tools for High Throughput RRNA Analysis. Nucleic Acids Res. 2014, 42, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE Database for Molecular Identification of Fungi: Handling Dark Taxa and Parallel Taxonomic Classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Kim, B.R.; Shin, J.; Guevarra, R.; Lee, J.H.; Kim, D.W.; Seol, K.-H.; Lee, J.-H.; Kim, H.B.; Isaacson, R.E. Deciphering Diversity Indices for Better Understanding of the Microbial Communities. J. Microbiol. Biotechnol. 2017, 27, 2089–2093. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. 2020. Available online: Https://Github.Com/Vegandevs/Vegan (accessed on 1 November 2021).
- Auffret, M.D.; Dewhurst, R.J.; Duthie, C.; Rooke, J.A.; Wallace, R.J.; Freeman, T.C.; Stewart, R.; Watson, M.; Roehe, R. The Rumen Microbiome as a Reservoir of Antimicrobial Resistance and Pathogenicity Genes Is Directly Affected by Diet in Beef Cattle. Microbiome 2017, 5, 159. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: Https://Www.R-Project.Org/ (accessed on 1 November 2021).
- Tomkins, N.W.; Denman, S.E.; Pilajun, R.; Wanapat, M.; Mcsweeney, C.S.; Elliott, R. Manipulating Rumen Fermentation and Methanogenesis Using an Essential Oil and Monensin in Beef Cattle Fed a Tropical Grass Hay. Anim. Feed Sci. Technol. 2015, 200, 25–34. [Google Scholar] [CrossRef]
- Torres, R.N.S.; Moura, D.C.; Ghedini, C.P.; Ezequiel, J.M.B.; Almeida, M.T.C. Meta-Analysis of the Effects of Essential Oils on Ruminal Fermentation and Performance of Sheep. Small Rumin. Res. 2020, 189, 106148. [Google Scholar] [CrossRef]
- Nocek, J.E.; Kautz, W.P.; Leedle, J.A.Z.; Block, E. Direct-Fed Microbial Supplementation on the Performance of Dairy Cattle during the Transition Period. J. Dairy Sci. 2003, 86, 331–335. [Google Scholar] [CrossRef] [PubMed]
- Belanche, A.; Doreau, M.; Edwards, J.E.; Moorby, J.M.; Pinloche, E.; Newbold, C.J. Shifts in the Rumen Microbiota Due to the Type of Carbohydrate and Level of Protein Ingested by Dairy Cattle Are Associated with Changes in Rumen Fermentation. J. Nutr. 2012, 142, 1684–1692. [Google Scholar] [CrossRef] [PubMed]
- Theodorou, M.K.; Mennim, G.; Davies, D.R.; Zhu, W.-Y.; Trinci, A.P.J.; Brookman, J.L. Anaerobic Fungi in the Digestive Tract of Mammalian Herbivores and Their Potential for Exploitation. Proc. Nutr. Soc. 1996, 55, 913–926. [Google Scholar] [CrossRef] [PubMed]
- Weimer, P.J.; Stevenson, D.M.; Mertens, D.R.; Thomas, E.E. Effect of Monensin Feeding and Withdrawal on Populations of Individual Bacterial Species in the Rumen of Lactating Dairy Cows Fed High-Starch Rations. Appl. Microbiol. Biotechnol. 2008, 80, 135–145. [Google Scholar] [CrossRef]
- Ben Shabat, S.K.; Sasson, G.; Doron-Faigenboim, A.; Durman, T.; Yaacoby, S.; Miller, M.E.B.; White, B.A.; Shterzer, N.; Mizrahi, I. Specific Microbiome-Dependent Mechanisms Underlie the Energy Harvest Efficiency of Ruminants. ISME J. 2016, 10, 2958–2972. [Google Scholar] [CrossRef] [PubMed]
- Lima, J.R.; Ribon, A.D.O.B.; Russell, J.B.; Mantovani, H.C. Bovicin HC5 Inhibits Wasteful Amino Acid Degradation by Mixed Ruminal Bacteria in Vitro. FEMS Microbiol. Lett. 2009, 292, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Ransom-Jones, E.; Jones, D.L.; McCarthy, A.J.; McDonald, J.E. The Fibrobacteres: An Important Phylum of Cellulose-Degrading Bacteria. Microb. Ecol. 2012, 63, 267–281. [Google Scholar] [CrossRef]
- Ogunade, I.; Schweickart, H.; Andries, K.; Lay, J.; Adeyemi, J. Monensin Alters the Functional and Metabolomic Profile of Rumen Microbiota in Beef Cattle. Animals 2018, 8, 211. [Google Scholar] [CrossRef]
- Busquet, M.; Calsamiglia, S.; Ferret, A.; Cardozo, P.W.; Kamel, C. Effects of Cinnamaldehyde and Garlic Oil on Rumen Microbial Fermentation in a Dual Flow Continuous Culture. J. Dairy Sci. 2005, 88, 2508–2516. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, F.; Chen, Y.; Wu, H.; Meng, Q. Metatranscriptomic Profiling Reveals the Effect of Breed on Active Rumen Eukaryotic Composition in Beef Cattle with Varied Feed Efficiency. Front. Microbiol. 2020, 11, 367. [Google Scholar] [CrossRef]
- Elliott, R.; Ash, A.J.; Calderon-Cortes, F.; Norton, B.W.; Bauchop, T. The Influence of Anaerobic Fungi on Rumen Volatile Fatty Acid Concentrations in Vivo. J. Agric. Sci. 1987, 109, 13–17. [Google Scholar] [CrossRef]
- Eren, A.M.; Sogin, M.L.; Morrison, H.G.; Vineis, J.H.; Fisher, J.C.; Newton, R.J.; Mclellan, S.L. A Single Genus in the Gut Microbiome Reflects Host Preference and Specificity. ISME J. 2015, 9, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Sanabria, E.; Guan, L.L.; Goonewardene, L.A.; Li, M.; Mujibi, D.F.; Stothard, P.; Moore, S.S.; Leon-Quintero, M.C. Correlation of Particular Bacterial PCR-Denaturing Gradient Gel Electrophoresis Patterns with Bovine Ruminal Fermentation Parameters and Feed Efficiency Traits. Appl. Environ. Microbiol. 2010, 76, 6338–6350. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhou, M.; Ominski, K.; Guan, L.L. Does the Rumen Microbiome Play a Role in Feed Efficiency of Beef Cattle? J. Anim. Sci. 2016, 94 (Suppl. S6), 44–49. [Google Scholar] [CrossRef]
- Li, F.; Guan, L.L. Metatranscriptomic Profiling Reveals Linkages between the Active Rumen Microbiome and Feed Efficiency in Beef Cattle. Appl. Environ. Microbiol. 2017, 83, e00061-17. [Google Scholar] [CrossRef]
- Paz, H.A.; Hales, K.E.; Wells, J.E.; Kuehn, L.A.; Freetly, H.C.; Berry, E.D.; Flythe, M.D.; Spangler, M.L.; Fernando, S.C. Rumen Bacterial Community Structure Impacts Feed Efficiency in Beef Cattle. J. Anim. Sci. 2018, 96, 1045–1058. [Google Scholar] [CrossRef]
- McGovern, E.; Kenny, D.A.; McCabe, M.S.; Fitzsimons, C.; McGee, M.; Kelly, A.K.; Waters, S.M. 16S RRNA Sequencing Reveals Relationship between Potent Cellulolytic Genera and Feed Efficiency in the Rumen of Bulls. Front. Microbiol. 2018, 9, 1842. [Google Scholar] [CrossRef]
- Meehan, C.J.; Beiko, R.G. A Phylogenomic View of Ecological Specialization in the Lachnospiraceae, a Family of Digestive Tract-Associated Bacteria. Genome Biol. Evol. 2014, 6, 703–713. [Google Scholar] [CrossRef]
- Guan, L.L.; Nkrumah, J.D.; Basarab, J.A.; Moore, S.S. Linkage of Microbial Ecology to Phenotype: Correlation of Rumen Microbial Ecology to Cattle’s Feed Efficiency. FEMS Microbiol. Lett. 2008, 288, 85–91. [Google Scholar] [CrossRef]
- Yang, W.Z.; Ametaj, B.N.; Benchaar, C.; Beauchemin, K.A. Dose Response to Cinnamaldehyde Supplementation in Growing Beef Heifers: Ruminal and Intestinal Digestion. J. Anim. Sci. 2010, 88, 680–688. [Google Scholar] [CrossRef]
- Benchaar, C.; Calsamiglia, S.; Chaves, A.V.; Fraser, G.R.; Colombatto, D.; Mcallister, T.A.; Beauchemin, K.A. A Review of Plant-Derived Essential Oils in Ruminant Nutrition and Production. Anim. Feed Sci. Technol. 2008, 145, 209–228. [Google Scholar] [CrossRef]
- Petri, R.M.; Schwaiger, T.; Penner, G.B.; Beauchemin, K.A.; Forster, R.J.; Mckinnon, J.J.; Mcallister, T.A. Characterization of the Core Rumen Microbiome in Cattle during Transition from Forage to Concentrate as Well as during and after an Acidotic Challenge. PLoS ONE 2013, 8, e83424. [Google Scholar] [CrossRef] [PubMed]
- Riddell, J.B.; Mcleod, K.R.; Cv, S.A. De Addition of a Bacillus Based Probiotic to the Diet of Preruminant Calves: Influence on Growth, Health, and Blood Parameters. Int. J. Appl. Res. Vet. Med. 2010, 8, 78–85. [Google Scholar]
- Shin, N.; Whon, T.W.; Bae, J. Proteobacteria: Microbial Signature of Dysbiosis in Gut Microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Eschenlauer, S.C.P.; McKain, N.; Walker, N.D.; McEwan, N.R.; Newbold, C.J.; Wallace, R.J. Ammonia Production by Ruminal Microorganisms and Enumeration, Isolation and Characterization of Bacteria Capable of Growth on Peptides and Amino Acids from Sheep Rumen. Appl. Environ. Microbiol. 2002, 68, 4925–4931. [Google Scholar] [CrossRef]
- Henderson, G.; Cox, F.; Ganesh, S.; Jonker, A.; Young, W.; Collaborators, G.R.C.; Janssen, P.H. Rumen Microbial Community Composition Varies with Diet and Host, but a Core Microbiome Is Found across a Wide Geographical Range. Sci. Rep. 2015, 5, 14567. [Google Scholar] [CrossRef]
- O’Hara, E.; Kelly, A.; McCabe, M.S.; Kenny, D.A.; Guan, L.L.; Waters, S.M. Effect of a Butyrate-Fortified Milk Replacer on Gastrointestinal Microbiota and Products of Fermentation in Artificially Reared Dairy Calves at Weaning. Sci. Rep. 2018, 8, 14901. [Google Scholar] [CrossRef]
- Gruninger, R.J.; Puniya, A.K.; Callaghan, T.M.; Edwards, J.E.; Youssef, N.; Dagar, S.; Fliegerova, K.; Griffith, G.W.; Forster, R.; Tsang, A.; et al. Anaerobic Fungi (Phylum Neocallimastigomycota): Advances in Understanding Their Taxonomy, Life Cycle, Ecology, Role and Biotechnological Potential. FEMS Microbiol. Ecol. 2014, 90, 1–17. [Google Scholar] [CrossRef]
- Han, X.; Li, B.; Wang, X.; Chen, Y.; Yang, Y. Effect of Dietary Concentrate to Forage Ratios on Ruminal Bacterial and Anaerobic Fungal Populations of Cashmere Goats. Anaerobe 2019, 59, 118–125. [Google Scholar] [CrossRef]

| Performance Traits | CON | MON | PRO | EO | p-Value |
|---|---|---|---|---|---|
| LW (kg) | 468 ± 26 | 471 ± 27 | 455 ± 34 | 460 ± 30 | 0.497 |
| ADG (kg/day) | 1.81 ± 0.11 | 1.85 ± 0.08 | 1.70 ± 0.71 | 1.68 ± 0.16 | 0.603 |
| DFI (kg/day) | 11.30 ± 0.38 a | 11.60 ± 0.71 a | 10.50 ± 0.43 b | 11.60 ± 0.47 a | 0.037 * |
| FCR | 6.25 ± 0.52 | 6.25 ± 0.46 | 6.24 ± 0.66 | 6.91 ± 0.46 | 0.255 |
| CON | MON | PRO | EO | p-Value | |
|---|---|---|---|---|---|
| Starter | |||||
| tVFA | 70.07 ± 23.17 | 84.85 ± 18.87 | 73.03 ± 18.03 | 81.65 ± 13.27 | 0.589 |
| Acetate | 66.33 ± 2.42 a | 57.99 ± 1.54 b | 65.26 ± 2.58 a | 66.97 ± 2.94 a | 0.033 * |
| Propionate | 16.17 ± 4.86 a | 28.87 ± 3.38 b | 17.67 ± 2.44 a | 16.96 ± 2.72 a | 0.033 * |
| Butyrate | 12.70 ± 2.38 | 9.91 ± 1.70 | 12.73 ± 1.58 | 12.44 ± 1.96 | 0.277 |
| A:P | 4.52 ± 1.44 a | 2.04 ± 0.28 b | 3.77 ± 0.61 a | 4.08 ± 0.81 a | 0.034 * |
| Grower | |||||
| tVFA | 105.19 ± 26.02 | 110.48 ± 13.57 | 100.01 ± 15.74 | 91.20 ± 15.15 | 0.657 |
| Acetate | 58.63 ± 0.81 | 58.46 ± 2.57 | 57.75 ± 2.41 | 59.56 ± 2.43 | 0.724 |
| Propionate | 28.19 ± 2.30 | 26.84 ± 2.57 | 28.94 ± 3.12 | 22.43 ± 5.36 | 0.235 |
| Butyrate | 8.19 ± 1.32 | 9.73 ± 0.29 | 9.41 ± 0.31 | 13.16 ± 5.13 | 0.134 |
| A:P | 2.09 ± 0.17 | 2.21 ± 0.29 | 2.03 ± 0.31 | 2.84 ± 0.79 | 0.474 |
| Finisher | |||||
| tVFA | 110.59 ± 13.10 | 94.41 ± 11.23 | 115.10 ± 18.52 | 95.03 ± 18.72 | 0.231 |
| Acetate | 56.16 ± 1.07 | 59.65 ± 3.26 | 56.23 ± 1.53 | 55.96 ± 1.56 | 0.382 |
| Propionate | 29.59 ± 1.89 | 22.52 ± 6.37 | 29.51 ± 2.11 | 29.62 ± 1.91 | 0.531 |
| Butyrate | 8.95 ± 1.33 | 10.96 ± 2.29 | 9.29 ± 0.99 | 9.29 ± 1.43 | 0.562 |
| A:P | 1.91 ± 0.15 | 2.89 ± 0.86 | 1.92 ± 0.18 | 1.90 ± 0.16 | 0.171 |
| Alpha Diversity Indices | CON | MON | EO | PRO | p-Value |
|---|---|---|---|---|---|
| Starter | |||||
| Observed number of ASVs | 1398 ± 48 | 1213 ± 56 | 1277 ± 70 | 1186 ± 105 | 0.461 |
| Chao1 Index | 1402 ± 49 | 1222 ± 55 | 1281 ± 70 | 1196 ± 106 | 0.492 |
| Shannon Index | 6.03 ± 0.15 | 5.74 ± 0.09 | 5.88 ± 0.21 | 5.65 ± 0.21 | 0.576 |
| Grower | |||||
| Observed number of ASVs | 805 ± 53 ab | 969 ± 37 a | 808 ± 47 ab | 701 ± 33 b | 0.046 * |
| Chao1 Index | 816 ± 54 ab | 980 ± 39 a | 819 ± 48 ab | 708 ± 32 b | 0.046 * |
| Shannon Index | 3.92 ± 0.20 | 4.33 ± 0.12 | 4.39 ± 0.13 | 3.95 ± 0.19 | 0.306 |
| Finisher | |||||
| Observed number of ASVs | 626 ± 12 | 608 ± 17 | 641 ± 66 | 737 ± 20 | 0.100 |
| Chao1 Index | 629 ± 13 | 612 ± 17 | 649 ± 66 | 742 ± 20 | 0.108 |
| Shannon Index | 2.98 ± 0.13 a | 3.63 ± 0.21 ab | 3.43 ± 0.24 ab | 4.13 ± 0.11 b | 0.044 * |
| Alpha Diversity Indices | CON | MON | EO | PRO | p-Value |
|---|---|---|---|---|---|
| Starter | |||||
| Observed number of ASVs | 263 ± 12 | 275 ± 15 | 275 ± 10 | 284 ± 12 | 0.814 |
| Chao1 Index | 264 ± 12 | 276 ± 15 | 276 ± 10 | 285 ± 12 | 0.782 |
| Shannon Index | 3.52 ± 0.17 | 3.65 ± 0.07 | 3.69 ± 0.18 | 3.86 ± 0.12 | 0.405 |
| Grower | |||||
| Observed number of ASVs | 304 ± 12 a | 287 ± 14 ab | 293 ± 10 ab | 249 ± 5 b | 0.055 ** |
| Chao1 Index | 304 ± 12 a | 287 ± 13 ab | 294 ± 10 ab | 250 ± 5 b | 0.063 ** |
| Shannon Index | 4.10 ± 0.15 | 3.77 ± 0.31 | 4.27 ± 0.02 | 4.17 ± 0.02 | 0.362 |
| Finisher | |||||
| Observed number of ASVs | 175 ± 4 | 151 ± 8 | 149 ± 5 | 153 ± 10 | 0.186 |
| Chao1 Index | 175 ± 4 | 152 ± 9 | 149 ± 5 | 153 ± 10 | 0.183 |
| Shannon Index | 3.30 ± 0.05 | 3.17 ± 0.09 | 3.21 ± 0.06 | 3.15 ± 0.08 | 0.618 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Linde, D.A.; Schokker, D.; du Toit, C.J.L.; Ramkilawon, G.D.; van Marle-Köster, E. The Effect of a Bacillus Probiotic and Essential Oils Compared to an Ionophore on the Rumen Microbiome Composition of Feedlot Cattle. Animals 2023, 13, 2927. https://doi.org/10.3390/ani13182927
Linde DA, Schokker D, du Toit CJL, Ramkilawon GD, van Marle-Köster E. The Effect of a Bacillus Probiotic and Essential Oils Compared to an Ionophore on the Rumen Microbiome Composition of Feedlot Cattle. Animals. 2023; 13(18):2927. https://doi.org/10.3390/ani13182927
Chicago/Turabian StyleLinde, Dina A., Dirkjan Schokker, Cornelius J. L. du Toit, Gopika D. Ramkilawon, and Este van Marle-Köster. 2023. "The Effect of a Bacillus Probiotic and Essential Oils Compared to an Ionophore on the Rumen Microbiome Composition of Feedlot Cattle" Animals 13, no. 18: 2927. https://doi.org/10.3390/ani13182927





