Whole Transcriptome Sequencing Reveals miRNAs and ceRNA Networks in Duck Abdominal Fat Deposition
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Experimental Animals and Samples
2.3. Histological Studies
2.4. Total RNA Extraction and Sequencing
2.5. Data Processing and Analysis
2.6. Standard Data Analysis
2.7. miRNA Expression Profiles and Target Gene Prediction
2.8. The ceRNA Regulatory Network
2.9. GO and KEGG Functional Enrichment Analysis
2.10. Validation of Differential Genes and miRNAs
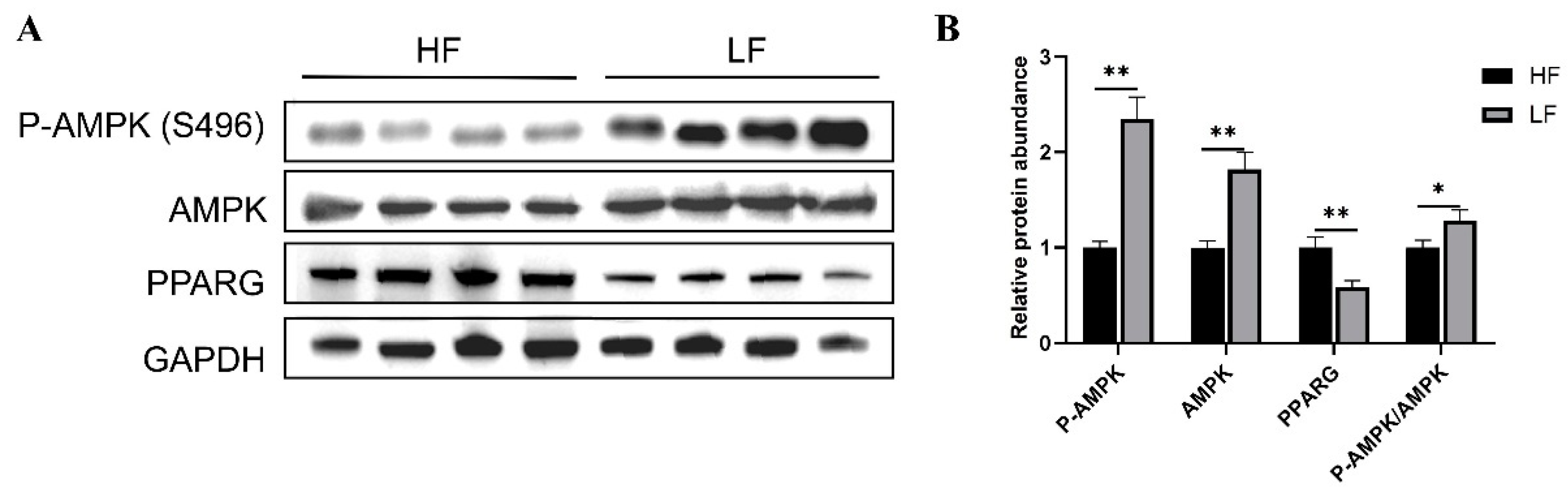
2.11. Western Blotting
2.12. Statistical Methods
3. Results
3.1. Histological Observation
3.2. Sequencing Data Quality Statistics
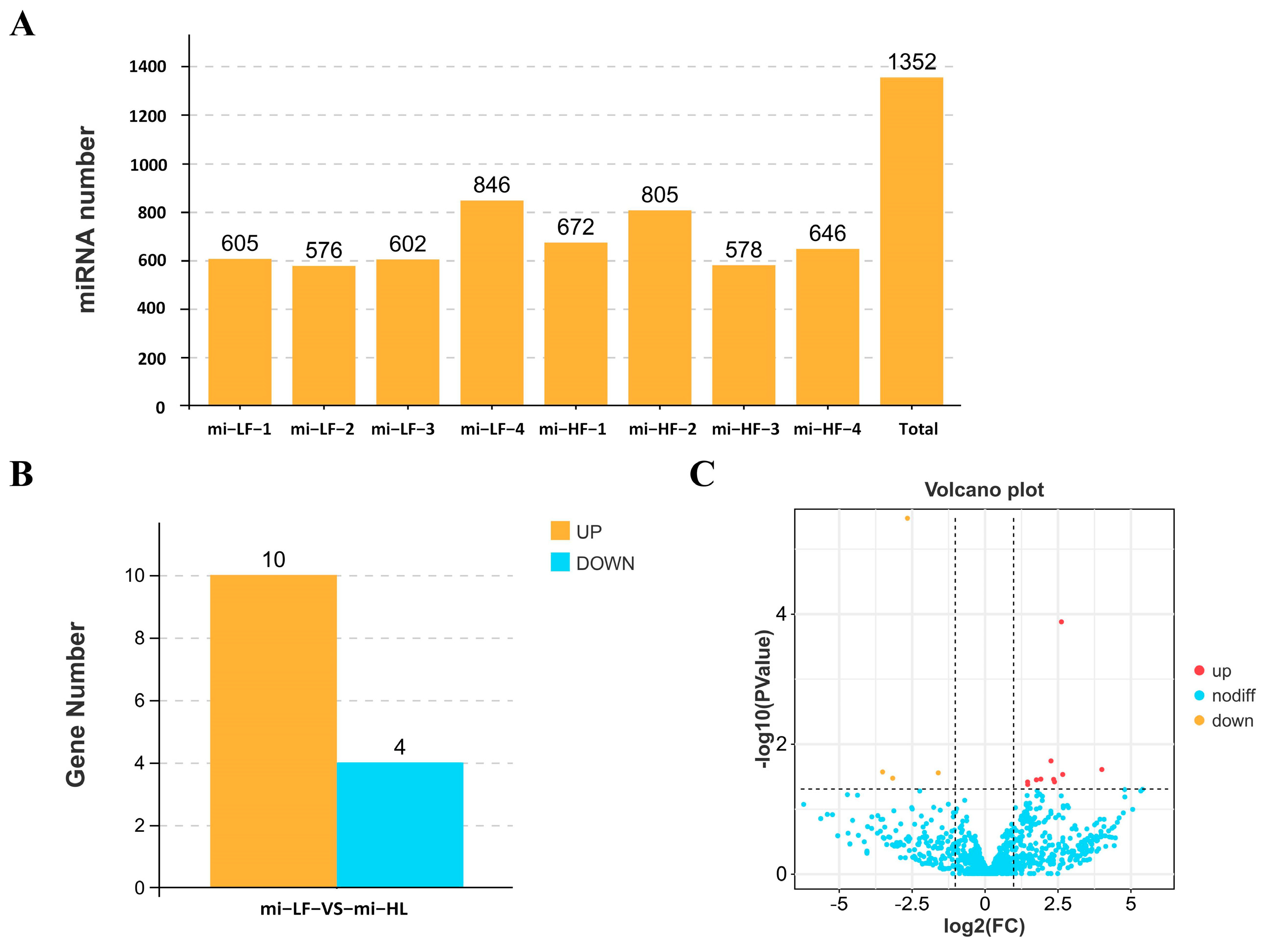
3.3. Identification and Differential Expression Analysis of miRNA
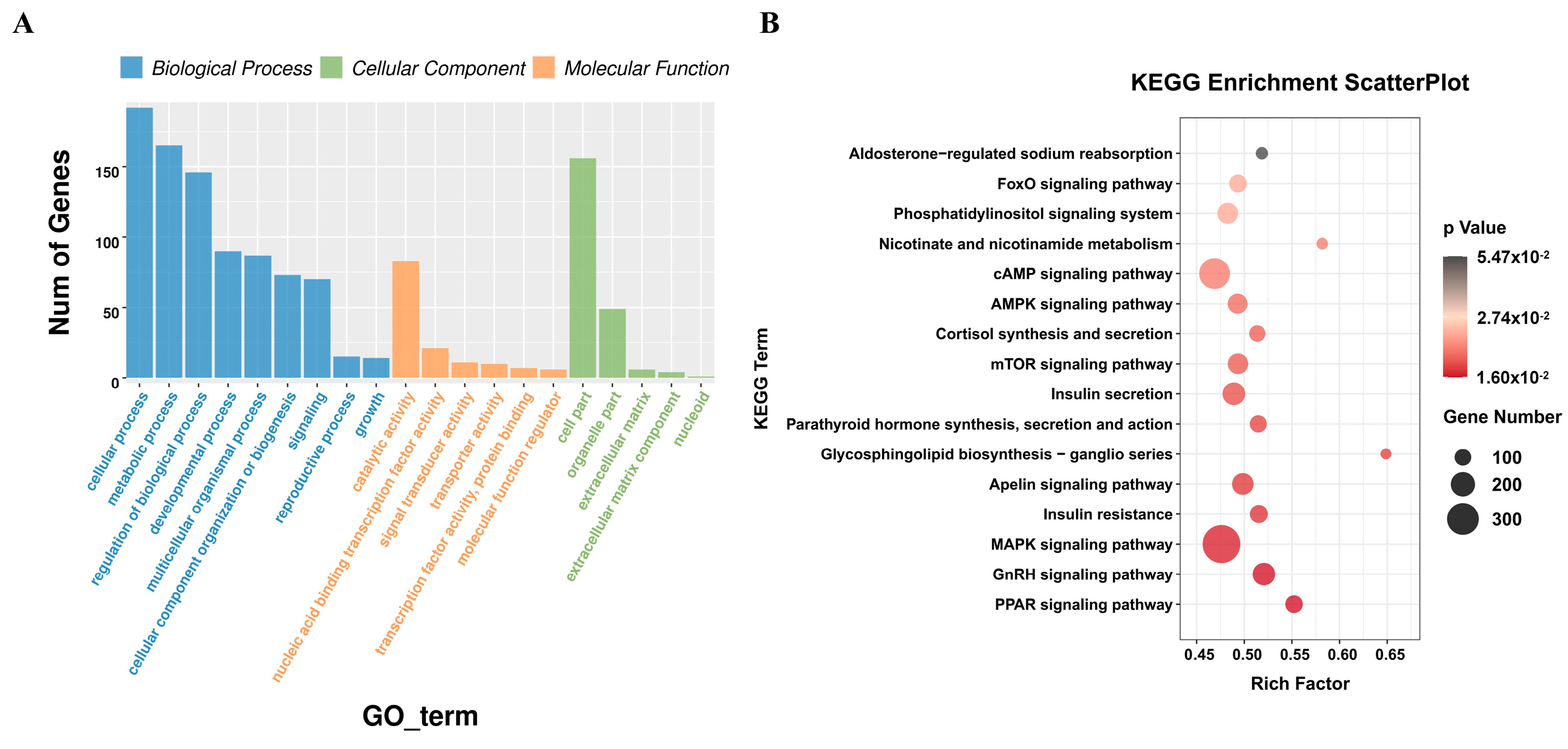
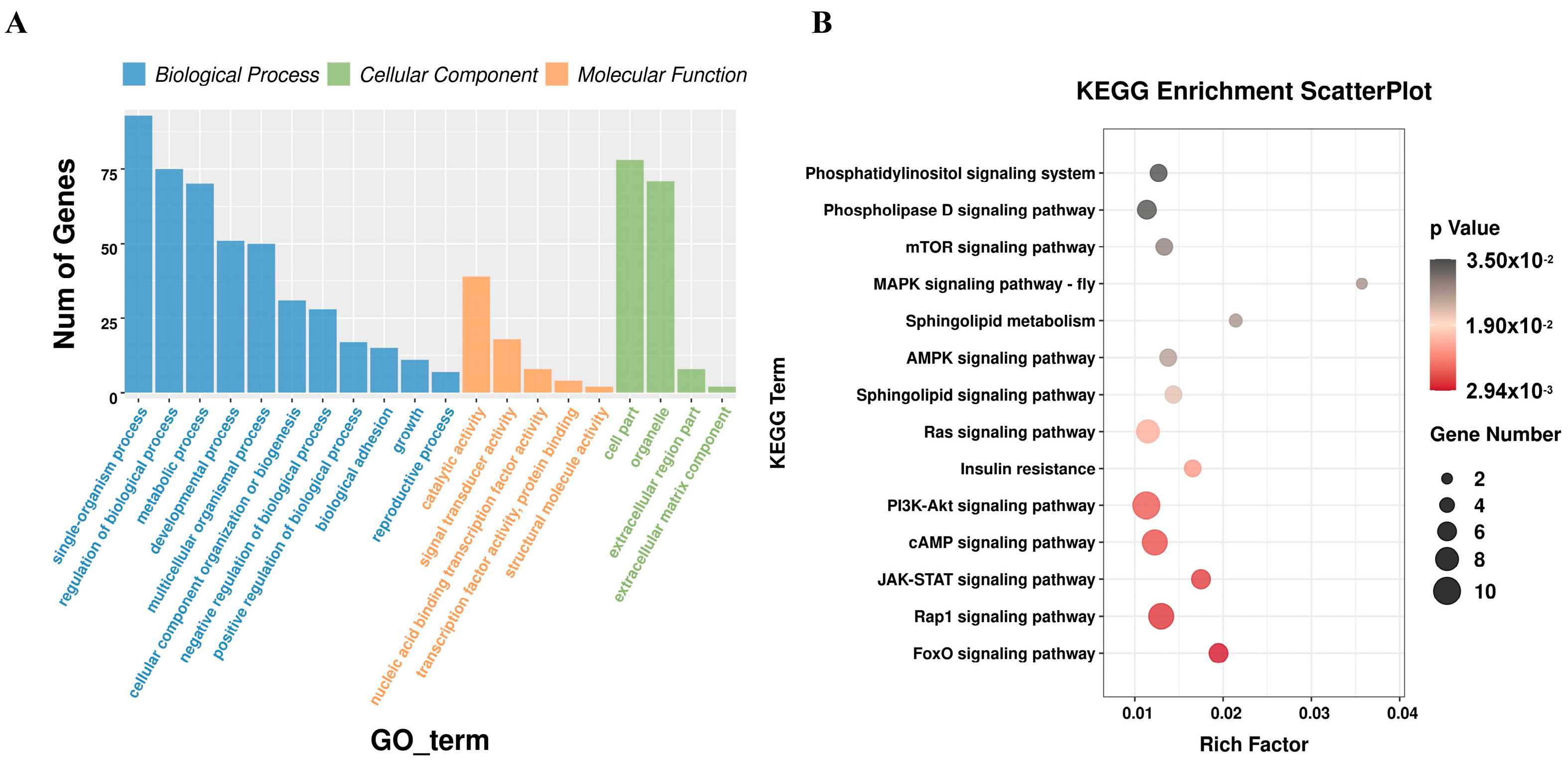
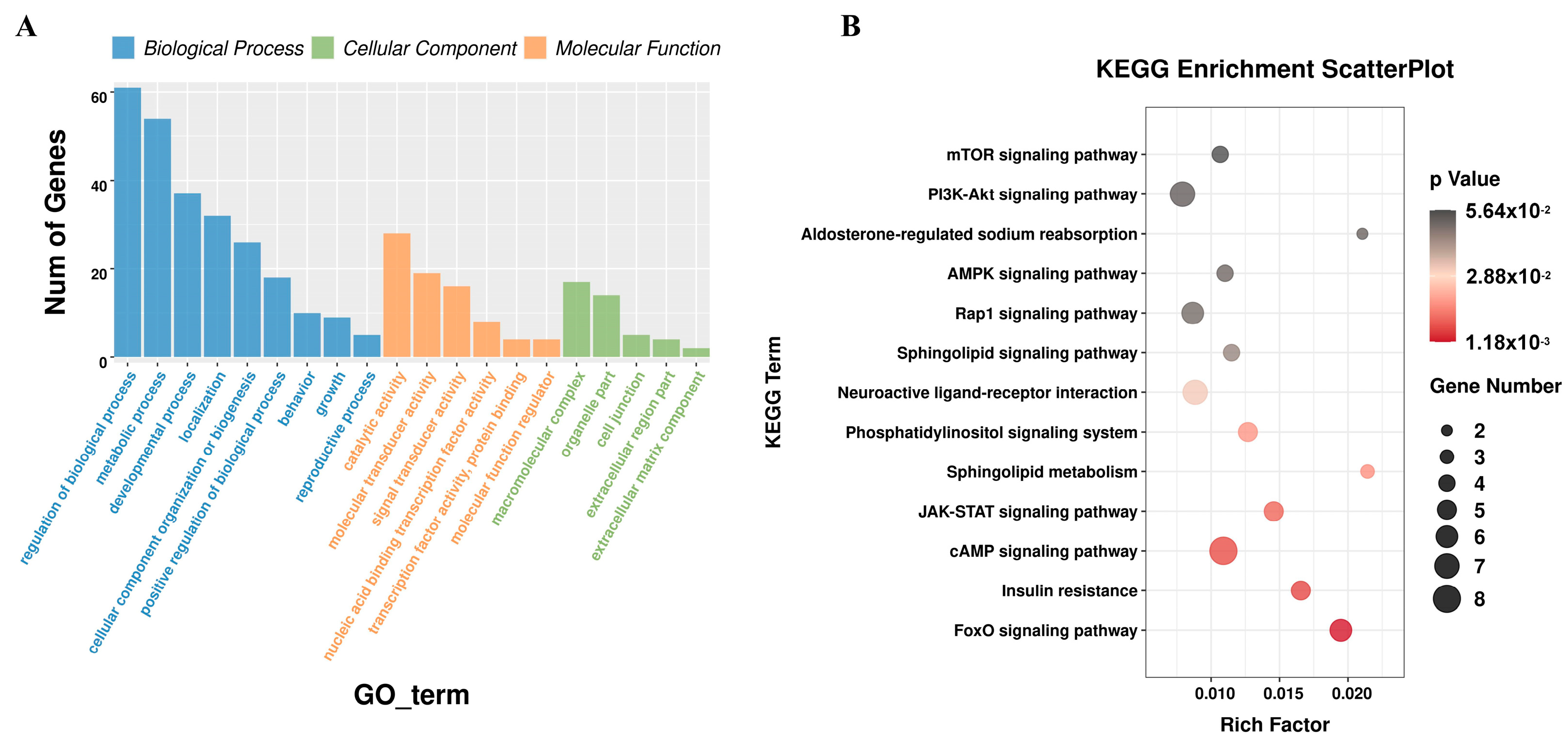
3.4. Functional Enrichment Analysis of miRNA Target Genes
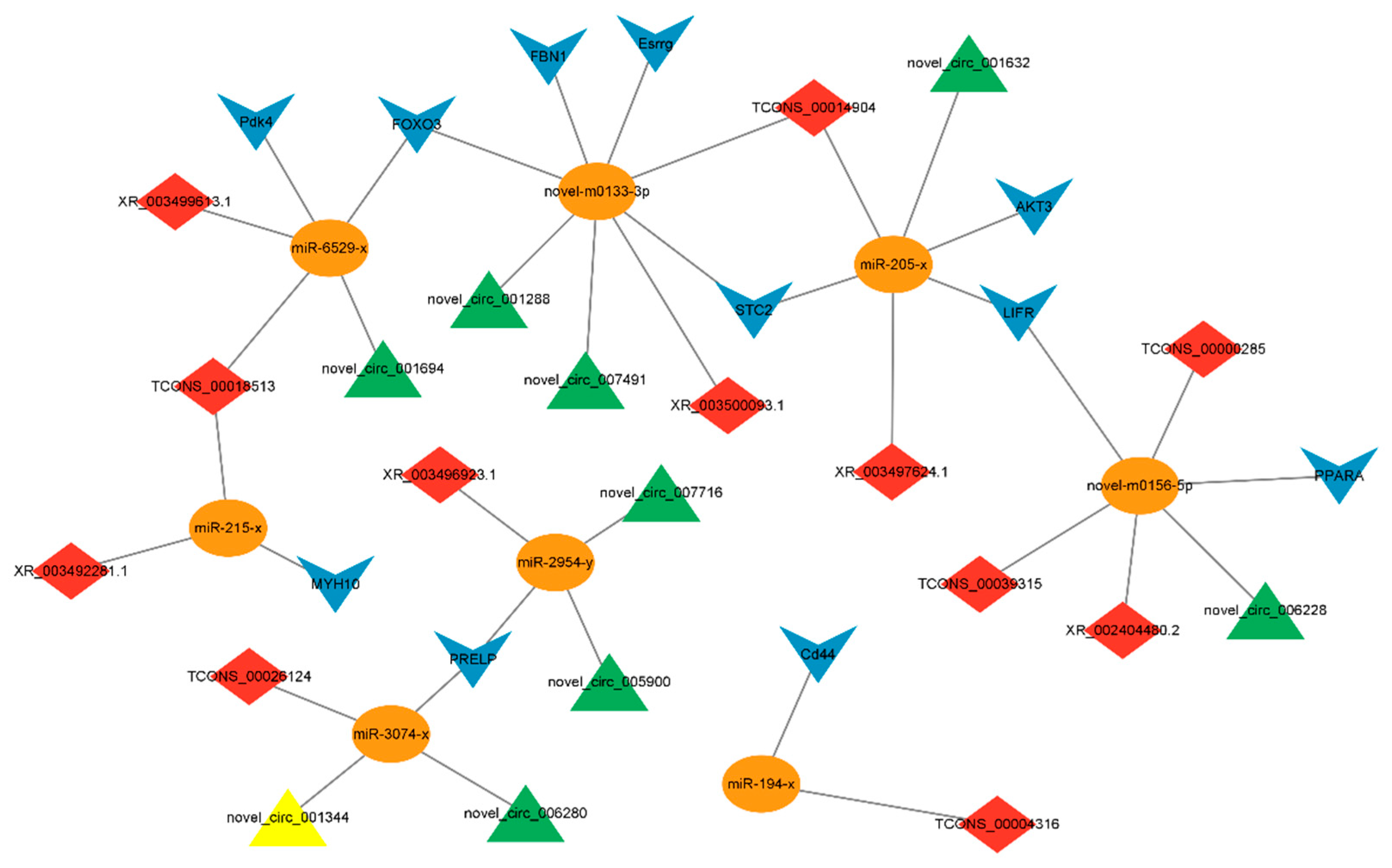
3.5. Construction and Analysis of ceRNAs Regulatory Network
3.6. Construction of Candidate ceRNA Network
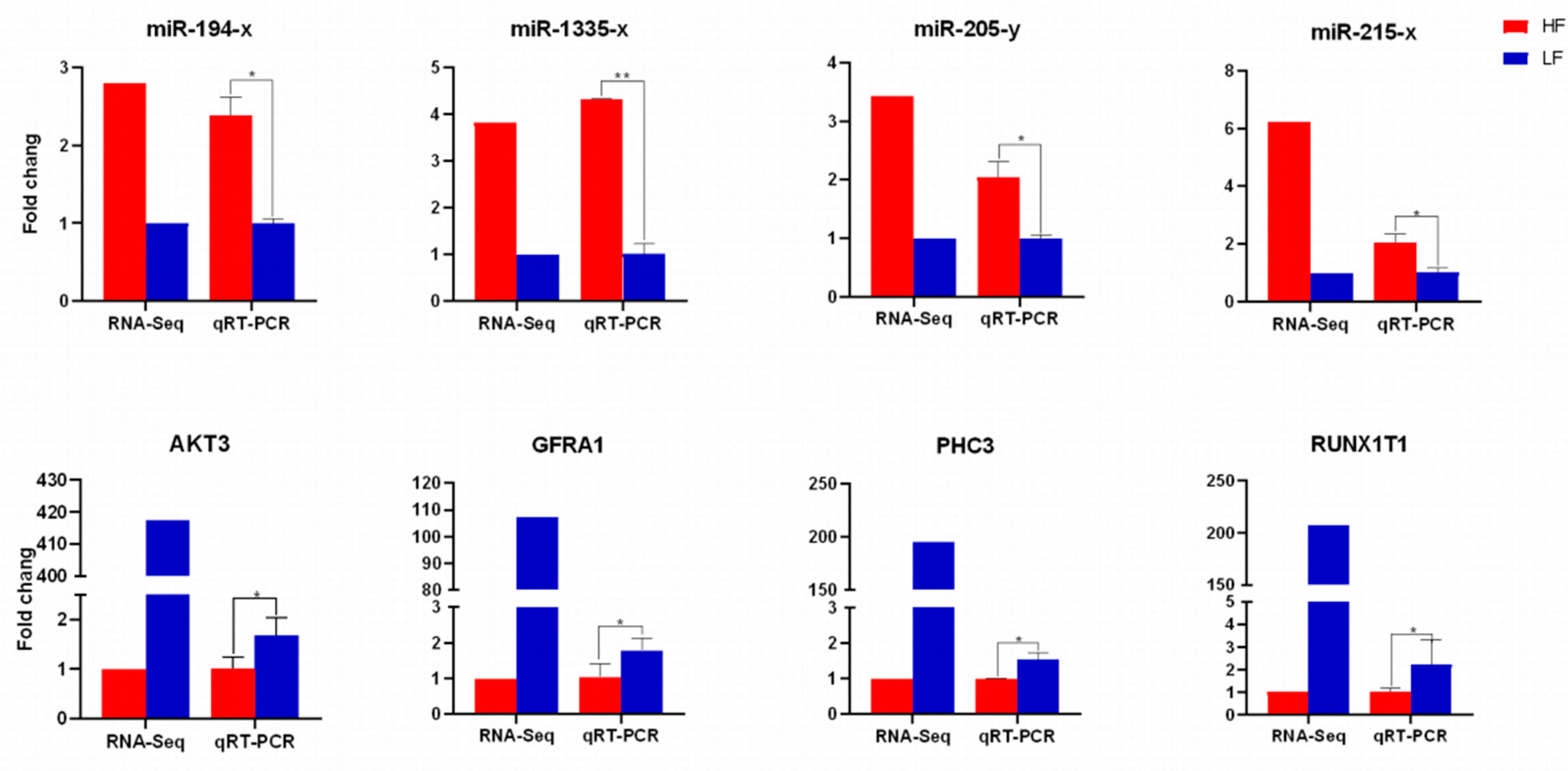
3.7. QRT-PCR and Western Blot Validation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ismail, I.; Joo, S.T. Poultry Meat Quality in Relation to Muscle Growth and Muscle Fiber Characteristics. Korean J. Food Sci. Anim. Resour. 2017, 37, 873–883. [Google Scholar] [PubMed]
- Yang, C.; Wang, Z.; Song, Q.; Dong, B.; Bi, Y.; Bai, H.; Jiang, Y.; Chang, G.; Chen, G. Transcriptome Sequencing to Identify Important Genes and lncRNAs Regulating Abdominal Fat Deposition in Ducks. Animals 2022, 12, 1256. [Google Scholar] [CrossRef]
- Lotfi, E.; Zerehdaran, S.; Ahani Azari, M. Genetic evaluation of carcass composition and fat deposition in Japanese quail. Poult. Sci. 2011, 90, 2202–2208. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Cai, M.; Zhu, J.; Xiao, W.; Liu, B.; Shi, Y.; Yang, X.; Liang, X.; Zheng, T.; Hu, S.; et al. miR-148a-3p promotes rabbit preadipocyte differentiation by targeting PTEN. In Vitro Cell. Dev. Biol. Anim. 2018, 54, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Yan, X.; Qiao, L.; Li, B.; Cheng, L.; Pan, Y.; Jing, J.; Cao, N.; Liu, W. MiR-330-5p negatively regulates ovine preadipocyte differentiation by targeting branched-chain aminotransferase 2. Anim. Sci. J. 2018, 89, 858–867. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Cai, B.; Jiang, L.; Abdalla, B.A.; Li, Z.; Nie, Q.; Zhang, X. lncRNA-Six1 Is a Target of miR-1611 that Functions as a ceRNA to Regulate Six1 Protein Expression and Fiber Type Switching in Chicken Myogenesis. Cells 2018, 7, 243. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Shi, Q.; Wang, S.; Zhang, H.; Xu, S. Ammonia regulates chicken tracheal cell necroptosis via the LncRNA-107053293/MiR-148a-3p/FAF1 axis. J. Hazard. Mater. 2020, 386, 121626. [Google Scholar] [CrossRef]
- Fu, S.; Zhao, Y.; Li, Y.; Li, G.; Chen, Y.; Li, Z.; Sun, G.; Li, H.; Kang, X.; Yan, F. Characterization of miRNA transcriptome profiles related to breast muscle development and intramuscular fat deposition in chickens. J. Cell. Biochem. 2018, 119, 7063–7079. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Wang, D.; Xia, Z.; Chen, C.; Cheng, P.; Xie, H.; Luo, X. The role of microRNAs in adipocyte differentiation. Front. Med. 2013, 7, 223–230. [Google Scholar] [CrossRef]
- Huang, C.J.; Choo, K.B.; Chen, C.F. The MicroRNA-signaling-peroxisome proliferator-activated receptor gamma connection in the modulation of adipogenesis: Bioinformatics projection on chicken. Poult. Sci. 2022, 101, 101950. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.Y.; Chae, M.K.; Lee, J.H.; Lee, E.J.; Yoon, J.S. MicroRNA-27 inhibits adipogenic differentiation in orbital fibroblasts from patients with Graves' orbitopathy. PLoS ONE 2019, 14, e0221077. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, S.; Cheng, G.; Mei, C.; Li, S.; Zhang, W.; Junjvlieke, Z.; Zan, L. MiR-145 reduces the activity of PI3K/Akt and MAPK signaling pathways and inhibits adipogenesis in bovine preadipocytes. Genomics 2020, 112, 2688–2694. [Google Scholar] [CrossRef]
- Zarkesh, M.; Tabaei, K.; Akbarzadeh, M.; Daneshafrooz, A.; Zadeh-Vakili, A. Association of miR-34a and miR-143 levels with PPARγ gene expression in adipose tissues of non-diabetic adults. J. Physiol. Anthropol. 2022, 41, 13. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Yuan, G.; He, J.; Shao, Y.; Zhang, J.; Guo, X. Aberrant expression of miR-214 is associated with obesity-induced insulin resistance as a biomarker and therapeutic. Diagn. Pathol. 2020, 15, 18. [Google Scholar] [CrossRef] [PubMed]
- Xi, F.X.; Wei, C.S.; Xu, Y.T.; Ma, L.; He, Y.L.; Shi, X.E.; Yang, G.S.; Yu, T.Y. MicroRNA-214-3p Targeting Ctnnb1 Promotes 3T3-L1 Preadipocyte Differentiation by Interfering with the Wnt/β-Catenin Signaling Pathway. Int. J. Mol. Sci. 2019, 20, 1816. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liang, W.; Wang, S.; Wang, Z.; Bai, H.; Jiang, Y.; Bi, Y.; Chen, G.; Chang, G. Circular RNA expression profiling reveals that circ-PLXNA1 functions in duck adipocyte differentiation. PLoS ONE 2020, 15, e0236069. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Zhai, B.; Zhao, Y.; Fan, S.; Yuan, P.; Li, H.; Li, S.; Li, Y.; Zhang, Y.; Huang, H.; Li, H.; et al. Differentially Expressed lncRNAs Related to the Development of Abdominal Fat in Gushi Chickens and Their Interaction Regulatory Network. Front. Genet. 2021, 12, 802857. [Google Scholar] [CrossRef]
- Xiao, C.; Sun, T.; Yang, Z.; Zou, L.; Deng, J.; Yang, X. Whole-transcriptome RNA sequencing reveals the global molecular responses and circRNA/lncRNA-miRNA-mRNA ceRNA regulatory network in chicken fat deposition. Poult. Sci. 2022, 101, 102121. [Google Scholar] [CrossRef]
- Wang, G.; Kim, W.K.; Cline, M.A.; Gilbert, E.R. Factors affecting adipose tissue development in chickens: A review. Poult. Sci. 2017, 96, 3687–3699. [Google Scholar] [CrossRef]
- Deng, K.; Ren, C.; Fan, Y.; Liu, Z.; Zhang, G.; Zhang, Y.; You, P.; Wang, F. miR-27a is an important adipogenesis regulator associated with differential lipid accumulation between intramuscular and subcutaneous adipose tissues of sheep. Domest. Anim. Endocrinol. 2020, 71, 106393. [Google Scholar] [CrossRef]
- Kurylowicz, A. microRNAs in Human Adipose Tissue Physiology and Dysfunction. Cells 2021, 10, 3342. [Google Scholar] [CrossRef]
- Sun, G.; Li, F.; Ma, X.; Sun, J.; Jiang, R.; Tian, Y.; Han, R.; Li, G.; Wang, Y.; Li, Z.; et al. gga-miRNA-18b-3p Inhibits Intramuscular Adipocytes Differentiation in Chicken by Targeting the ACOT13 Gene. Cells 2019, 8, 556. [Google Scholar] [CrossRef]
- Li, F.; Li, D.; Zhang, M.; Sun, J.; Li, W.; Jiang, R.; Han, R.; Wang, Y.; Tian, Y.; Kang, X.; et al. miRNA-223 targets the GPAM gene and regulates the differentiation of intramuscular adipocytes. Gene 2019, 685, 106–113. [Google Scholar] [CrossRef]
- Li, G.; Fu, S.; Chen, Y.; Jin, W.; Zhai, B.; Li, Y.; Sun, G.; Han, R.; Wang, Y.; Tian, Y.; et al. MicroRNA-15a Regulates the Differentiation of Intramuscular Preadipocytes by Targeting ACAA1, ACOX1 and SCP2 in Chickens. Int. J. Mol. Sci. 2019, 20, 4063. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Cheng, M.; Qiao, S.; Wang, Y.; Li, H.; Wang, N. Gga-miR-21 inhibits chicken pre-adipocyte proliferation in part by down-regulating Kruppel-like factor 5. Poult. Sci. 2017, 96, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Chen, Y.; Qin, L.; Cheng, L.; Ren, G.; Cong, P.; Mo, D.; He, Z. Effect of miR-205 on 3T3-L1 preadipocyte differentiation through targeting to glycogen synthase kinase 3 beta. Biotechnol. Lett. 2014, 36, 1233–1243. [Google Scholar] [CrossRef]
- John, E.; Wienecke-Baldacchino, A.; Liivrand, M.; Heinäniemi, M.; Carlberg, C.; Sinkkonen, L. Dataset integration identifies transcriptional regulation of microRNA genes by PPARγ in differentiating mouse 3T3-L1 adipocytes. Nucleic Acids Res. 2012, 40, 4446–4460. [Google Scholar] [CrossRef] [PubMed]
- Kubo, M.; Ijichi, N.; Ikeda, K.; Horie-Inoue, K.; Takeda, S.; Inoue, S. Modulation of adipogenesis-related gene expression by estrogen-related receptor gamma during adipocytic differentiation. Biochim. Biophys. Acta. 2009, 1789, 71–77. [Google Scholar]
- Wu, Z.; Cheng, H.; Liu, J.; Zhang, S.; Zhang, M.; Liu, F.; Li, Y.; Huang, Q.; Jiang, Y.; Chen, S.; et al. The Oncogenic and Diagnostic Potential of Stanniocalcin 2 in Hepatocellular Carcinoma. J. Hepatocell. Carcinoma 2022, 9, 141–155. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhou, X.; Fang, Z.; Pan, Z. Effect of STC2 gene silencing on colorectal cancer cells. Mol. Med. Rep. 2019, 20, 977–984. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Guo, C.; Chen, C.; Zhang, Y. Characterization of cellular senescence patterns predicts the prognosis and therapeutic response of hepatocellular carcinoma. Front. Mol. Biosci. 2022, 9, 1100285. [Google Scholar] [CrossRef]
- Li, S.; Huang, Q.; Li, D.; Lv, L.; Li, Y.; Wu, Z. The significance of Stanniocalcin 2 in malignancies and mechanisms. Bioengineered 2021, 12, 7276–7285. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Jia, Q.; Xing, Y.; Ma, C.; Guan, H.; Tian, W.; Kang, X.; Tian, Y.; Liu, X.; Li, H. STC2 Inhibits Hepatic Lipid Synthesis and Correlates with Intramuscular Fatty Acid Composition, Body Weight and Carcass Traits in Chickens. Animals 2024, 14, 383. [Google Scholar] [CrossRef] [PubMed]
- Manning, B.D.; Cantley, L.C. AKT/PKB signaling: Navigating downstream. Cell 2007, 129, 1261–1274. [Google Scholar] [CrossRef]
- Humphrey, S.J.; Yang, G.; Yang, P.; Fazakerley, D.J.; Stöckli, J.; Yang, J.Y.; James, D.E. Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab. 2013, 17, 1009–1020. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Zhang, L.; Biswas, S.; Schugar, R.C.; Brown, J.M.; Byzova, T.; Podrez, E. Akt3 inhibits adipogenesis and protects from diet-induced obesity via WNK1/SGK1 signaling. JCI Insight 2017, 2, e95687. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.R.; Jung, H.S.; Bae, S.W.; Kim, J.H.; Park, B.L.; Choi, Y.H.; Cho, H.Y.; Cheong, H.S.; Shin, H.D. Polymorphisms in FOXO gene family and association analysis with BMI. Obesity 2006, 14, 188–193. [Google Scholar] [CrossRef]
- Wan, Z.; Liu, X.; Yang, X.; Huang, Z.; Chen, X.; Feng, Q.; Cao, H.; Deng, H. MicroRNA-411-5p alleviates lipid deposition in metabolic dysfunction-associated steatotic liver disease by targeting the EIF4G2/FOXO3 axis. Cell. Mol. Life Sci. 2024, 81, 398. [Google Scholar] [CrossRef]
- Guo, T.; Gupta, A.; Yu, J.; Granados, J.Z.; Gandhi, A.Y.; Evers, B.M.; Iyengar, P.; Infante, R.E. LIFR-α-dependent adipocyte signaling in obesity limits adipose expansion contributing to fatty liver disease. iScience 2021, 24, 102227. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, Y.; Yan, R.; Xu, X.; Gao, L.; Mei, J.; Liu, J.; Wang, X.; Zhang, J.; Wu, P.; et al. miR-377-3p regulates adipogenic differentiation of human bone marrow mesenchymal stem cells by regulating LIFR. Mol. Cell. Biochem. 2018, 449, 295–303. [Google Scholar] [CrossRef]
- Holness, M.J.; Sugden, M.C. Regulation of pyruvate dehydrogenase complex activity by reversible phosphorylation. Biochem. Soc. Trans. 2003, 31, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, I.K.N.; Tusubira, D.; Ashrafi, H.; Dyrstad, S.E.; Hansen, L.; Liu, X.Z.; Nilsson, L.I.H.; Løvsletten, N.G.; Berge, K.; Wergedahl, H.; et al. Upregulated PDK4 expression is a sensitive marker of increased fatty acid oxidation. Mitochondrion 2019, 49, 97–110. [Google Scholar] [CrossRef]
- Roche, T.E.; Hiromasa, Y. Pyruvate dehydrogenase kinase regulatory mechanisms and inhibition in treating diabetes, heart ischemia, and cancer. Cell. Mol. Life Sci. 2007, 64, 830–849. [Google Scholar] [CrossRef] [PubMed]
- Bougarne, N.; Weyers, B.; Desmet, S.J.; Deckers, J.; Ray, D.W.; Staels, B.; De Bosscher, K. Molecular Actions of PPARα in Lipid Metabolism and Inflammation. Endocr. Rev. 2018, 39, 760–802. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Shi, J.; Meng, Q.; Tang, Z.; Liu, T.; Zhang, Q.; Cheng, S. Verification of Key Target Molecules for Intramuscular Fat Deposition and Screening of SNP Sites in Sheep from Small-Tail Han Sheep Breed and Its Cross with Suffolk. Int. J. Mol. Sci. 2024, 25, 2951. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhang, A.; Liu, X.; Yang, Y.; Zhao, R.; Jia, Y. m6A-Mediated PPARA Translational Suppression Contributes to Corticosterone-Induced Visceral Fat Deposition in Chickens. Int. J. Mol. Sci. 2022, 23, 15761. [Google Scholar] [CrossRef] [PubMed]
- Goldblatt, J.; Hyatt, J.; Edwards, C.; Walpole, I. Further evidence for a marfanoid syndrome with neonatal progeroid features and severe generalized lipodystrophy due to frameshift mutations near the 3' end of the FBN1 gene. Am. J. Med. Genet. A 2011, 155A, 717–720. [Google Scholar] [CrossRef]
- Romere, C.; Duerrschmid, C.; Bournat, J.; Constable, P.; Jain, M.; Xia, F.; Saha, P.K.; Del Solar, M.; Zhu, B.; York, B.; et al. Asprosin, a Fasting-Induced Glucogenic Protein Hormone. Cell 2016, 165, 566–579. [Google Scholar] [CrossRef]
- Davis, M.R.; Arner, E.; Duffy, C.R.; De Sousa, P.A.; Dahlman, I.; Arner, P.; Summers, K.M. Expression of FBN1 during adipogenesis: Relevance to the lipodystrophy phenotype in Marfan syndrome and related conditions. Mol. Genet. Metab. 2016, 119, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Kislev, N.; Mor-Yossef Moldovan, L.; Barak, R.; Egozi, M.; Benayahu, D. MYH10 Governs Adipocyte Function and Adipogenesis through Its Interaction with GLUT4. Int. J. Mol. Sci. 2022, 23, 2367. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.S.; Liao, G.; DeGraff, L.M.; Gerrish, K.; Bortner, C.D.; Garantziotis, S.; Jetten, A.M. CD44 plays a critical role in regulating diet-induced adipose inflammation, hepatic steatosis, and insulin resistance. PLoS ONE 2013, 8, e58417. [Google Scholar] [CrossRef]
- Weng, X.; Jiang, H.; Walker, D.J.; Zhou, H.; Lin, D.; Wang, J.; Kang, L. Deletion of CD44 promotes adipogenesis by regulating PPARγ and cell cycle-related pathways. J. Endocrinol. 2024, 262, e240079. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Zheng, P.; Yan, X.Y.; Chen, H.J.; Fang, H.T.; Luo, Y.Y.; Peng, Y.X.; Zhang, L.; Yan, Y.E. Adipocyte-secreted PRELP promotes adipocyte differentiation and adipose tissue fibrosis by binding with p75NTR to activate FAK/MAPK signaling. Int. J. Biol. Macromol. 2024, 279, 135376. [Google Scholar] [CrossRef] [PubMed]
- Veiga-Lopez, A.; Moeller, J.; Sreedharan, R.; Singer, K.; Lumeng, C.; Ye, W.; Pease, A.; Padmanabhan, V. Developmental programming: Interaction between prenatal BPA exposure and postnatal adiposity on metabolic variables in female sheep. Am. J. Physiol. Endocrinol. Metab. 2016, 310, E238–E247. [Google Scholar] [CrossRef]
- Deng, K.; Ren, C.; Liu, Z.; Gao, X.; Fan, Y.; Zhang, G.; Zhang, Y.; Ma, E.S.; Wang, F.; You, P. Characterization of RUNX1T1, an Adipogenesis Regulator in Ovine Preadipocyte Differentiation. Int. J. Mol. Sci. 2018, 19, 1300. [Google Scholar] [CrossRef] [PubMed]



| Genes/miRNAs | GenBank | Primer Sequences (5′ → 3′) | Product Length/bp | Annealing Temperature/°C |
|---|---|---|---|---|
| miR-194-5p | - | Reverse: GTCGTATCCAGTGCAGGGTCCG AGGTATTCGCACTGGATACGACTCCCTG F: TATTGCAAACTCCGTGGCCT R: AGTGCAGGGTCCGAGGTATT | 22 | 60 |
| miR-215-5p | - | Reverse: GTCGTATCCAGTGCAGGGTCCG AGGTATTCGCACTGGATACGACAGTCTG F: GCGCGATGACCTATGAATTGA R: AGTGCAGGGTCCGAGGTATT | 21 | 60 |
| miR-205-5p | - | Reverse: GTCGTATCCAGTGCAGGGTCCG AGGTATTCGCACTGGATACGACCAGATT F: GCCCTTCATTCCACCGG R: AGTGCAGGGTCCGAGGTATT | 22 | 60 |
| miR-28-3p | - | Reverse: GTCGTATCCAGTGCAGGGTCCG AGGTATTCGCACTGGATACGACTCCAGG F: CGCGCACTAGATTGTGAGCT R: AGTGCAGGGTCCGAGGTATT | 22 | 60 |
| AKT3 | XM_027453846 | F: GGTGACGTCGGGAGTTTTC R: CTGATGAGTCACCCCCGGA | 101 | 60 |
| GFRA1 | XM_021274105 | F: ATTCTTCCCACATCCTGGCATT R: TCAAGCAAGTCATTTCCCTGT | 149 | 60 |
| PHC3 | XM_027464250 | F: CAACTGTACCACAGGCAAGC R: CTGTGCAGGTGTAGGAGAGG | 125 | 60 |
| RUNX1T1 | XM_027452458 | F: CATTCCAGAACAGGAGGCAC R: CTTCTCAGTGCGATCAGGCAT | 105 | 60 |
| Antibodies | Cat No. | Source | Predicted Band Size | Dilution of WB |
|---|---|---|---|---|
| P-AMPK | ET1612-72 | HUABIO, Hangzhou, China | 64 kDa | 1:1000 |
| AMPK | 5831S | CST, Danvers, MA, USA | 62 kDa | 1:1000 |
| PPARG | ab178860 | abcam, Cambridge, UK | 58 kDa | 1:1000 |
| GAPDH | ab181602 | abcam, Cambridge, UK | 36 kDa | 1:5000 |
| id | clean_reads | high_quality | 3′adapter_null | smaller_than_18nt | polyA | low cutoff | clean_tags |
|---|---|---|---|---|---|---|---|
| mi-HF-1 | 16836125 | 99.32% | 0.38% | 12.67% | 0.0005% | 3.19% | 81.91% |
| mi-HF-2 | 30279412 | 99.39% | 0.33% | 11.42% | 0.0008% | 3.68% | 83.57% |
| mi-HF-3 | 14406324 | 99.60% | 0.20% | 8.64% | 0.0008% | 4.89% | 84.61% |
| mi-HF-4 | 14835442 | 99.40% | 0.36% | 13.57% | 0.0006% | 5.55% | 79.59% |
| mi-LF-1 | 12356483 | 99.73% | 0.15% | 18.60% | 0.0007% | 8.22% | 71.98% |
| mi-LF-2 | 15701630 | 99.22% | 0.28% | 2.27% | 0.0007% | 1.99% | 94.95% |
| mi-LF-3 | 15106435 | 99.41% | 0.17% | 4.88% | 0.0009% | 3.93% | 90.44% |
| mi-LF-4 | 54771598 | 99.26% | 0.15% | 2.50% | 0.0005% | 1.54% | 95.31% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Yang, C.; Dong, B.; Chen, A.; Song, Q.; Bai, H.; Jiang, Y.; Chang, G.; Chen, G. Whole Transcriptome Sequencing Reveals miRNAs and ceRNA Networks in Duck Abdominal Fat Deposition. Animals 2025, 15, 506. https://doi.org/10.3390/ani15040506
Wang Z, Yang C, Dong B, Chen A, Song Q, Bai H, Jiang Y, Chang G, Chen G. Whole Transcriptome Sequencing Reveals miRNAs and ceRNA Networks in Duck Abdominal Fat Deposition. Animals. 2025; 15(4):506. https://doi.org/10.3390/ani15040506
Chicago/Turabian StyleWang, Zhixiu, Chunyan Yang, Bingqiang Dong, Anqi Chen, Qianqian Song, Hao Bai, Yong Jiang, Guobin Chang, and Guohong Chen. 2025. "Whole Transcriptome Sequencing Reveals miRNAs and ceRNA Networks in Duck Abdominal Fat Deposition" Animals 15, no. 4: 506. https://doi.org/10.3390/ani15040506
APA StyleWang, Z., Yang, C., Dong, B., Chen, A., Song, Q., Bai, H., Jiang, Y., Chang, G., & Chen, G. (2025). Whole Transcriptome Sequencing Reveals miRNAs and ceRNA Networks in Duck Abdominal Fat Deposition. Animals, 15(4), 506. https://doi.org/10.3390/ani15040506






