Molecular Insights into Epigenetics and Cannabinoid Receptors
Abstract
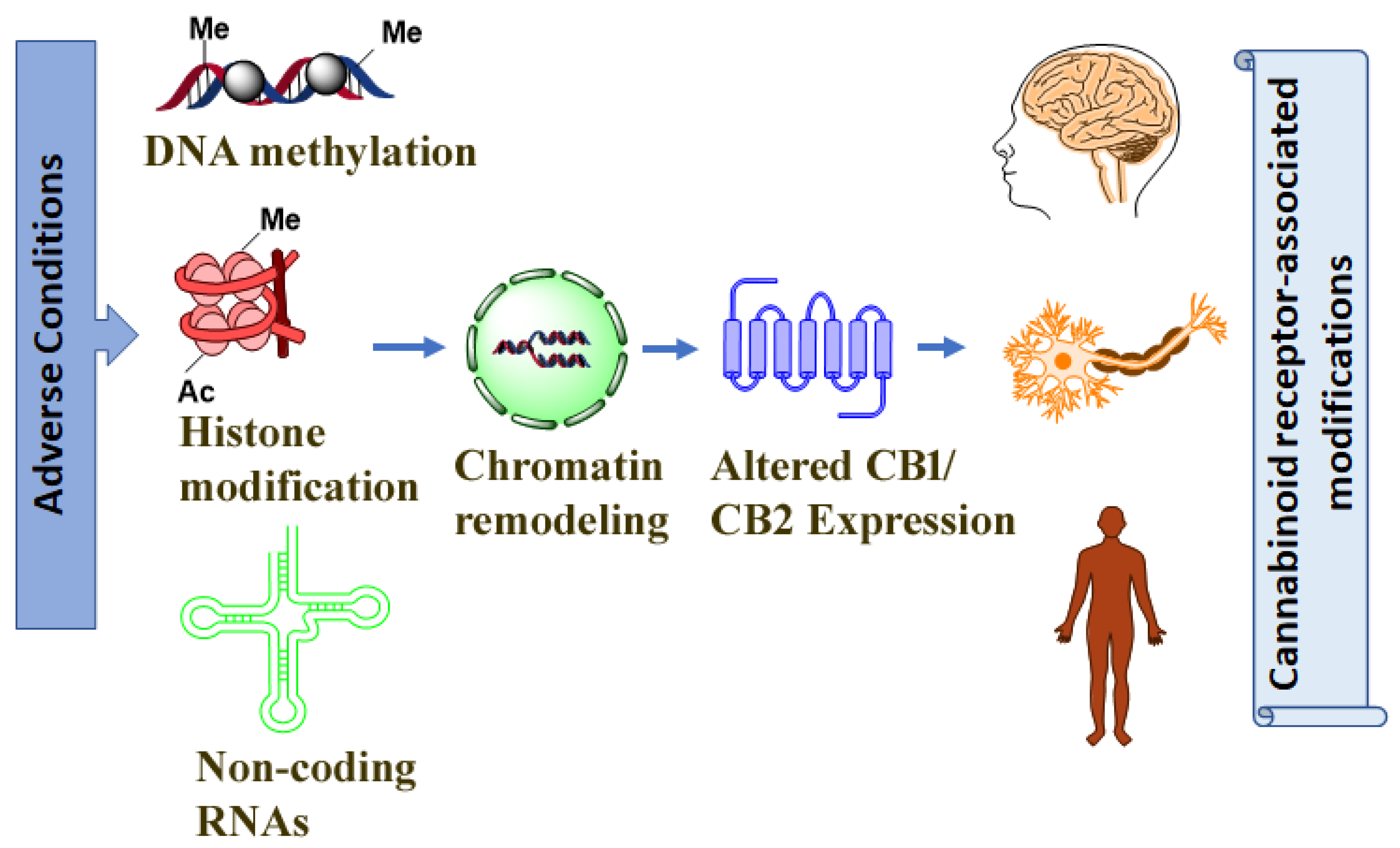
1. Introduction
1.1. Endocannabinoid System: A Brief Overview
1.2. Epigenetic Mechanisms: A Brief Overview
2. Role of DNA Methylation: Cannabinoid Receptors
2.1. DNA Methylation on CB1 Receptor Gene (Cnr1) Expression
2.2. DNA Methylation on CB2 Gene (Cnr2) Expression
2.3. Cannabinoid Receptor Stimulation on DNA Methylation
3. Role of Post-Translational Modification of DNA-Associated Histone Proteins: Cannabinoid Receptors
3.1. Histone Modifications That Modulate Cnr1 Gene Expression
3.2. Histone Modifications That Modulate Cnr2 Gene Expression
3.3. Modulation of Histone Acetylation and Methylation by CB1 and CB2 Activities
4. Regulation of microRNAs and Cannabinoid Receptors
4.1. miRNAs That Modulate Cnr1 Gene Expression
4.2. miRNAs That Modulate Cnr2 Gene Expression
4.3. Modulation of miRNAs by CB1 and CB2 Activities
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Devane, W.A.; Hanus, L.; Breuer, A.; Pertwee, R.G.; Stevenson, L.A.; Griffin, G.; Gibson, D.; Mandelbaum, A.; Etinger, A.; Mechoulam, R. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science 1992, 258, 1946–1949. [Google Scholar] [CrossRef] [PubMed]
- Di Marzo, V.; Fontana, A.; Cadas, H.; Schinelli, S.; Cimino, G.; Schwartz, J.-C.; Piomelli, D. Formation and inactivation of endogenous cannabinoid anandamide in central neurons. Nature 1994, 372, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Mechoulam, R.; Ben-Shabat, S.; Hanus, L.; Ligumsky, M.; Kaminski, N.E.; Schatz, A.R.; Gopher, A.; Almog, S.; Martin, B.R.; Compton, D.R.; et al. Identification of an endogenous 2-monoglyceride, present in canine gut, that binds to cannabinoid receptors. Biochem. Pharmacol. 1995, 50, 83–90. [Google Scholar] [CrossRef]
- Sugiura, T.; Kondo, S.; Sukagawa, A.; Nakane, S.; Shinoda, A.; Itoh, K.; Yamashita, A.; Waku, K. 2-Arachidonoylglycerol: A possible endogenous cannabinoid receptor ligand in brain. Biochem. Biophys. Res. Commun. 1995, 215, 89–97. [Google Scholar] [CrossRef]
- Okamoto, Y.; Morishita, J.; Tsuboi, K.; Tonai, T.; Ueda, N. Molecular Characterization of a Phospholipase D Generating Anandamide and Its Congeners. J. Biol. Chem. 2004, 279, 5298–5305. [Google Scholar] [CrossRef]
- Bisogno, T.; Howell, F.; Williams, G.; Minassi, A.; Cascio, M.G.; Ligresti, A.; Matias, I.; Schiano-Moriello, A.; Paul, P.; Williams, E.-J.; et al. Cloning of the first sn1-DAG lipases points to the spatial and temporal regulation of endocannabinoid signaling in the brain. J. Cell Biol. 2003, 163, 463–468. [Google Scholar] [CrossRef]
- Mechoulam, R. A Delightful Trip Along the Pathway of Cannabinoid and Endocannabinoid Chemistry and Pharmacology. Annu. Rev. Pharmacol. Toxicol. 2022, 63. [Google Scholar] [CrossRef]
- Veilleux, A.; Di Marzo, V.; Silvestri, C. The Expanded Endocannabinoid System/Endocannabinoidome as a Potential Target for Treating Diabetes Mellitus. Curr. Diabetes Rep. 2019, 19, 117. [Google Scholar] [CrossRef]
- Cravatt, B.F.; Giang, D.K.; Mayfield, S.P.; Boger, D.L.; Lerner, R.A.; Gilula, N.B. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature 1996, 384, 83–87. [Google Scholar] [CrossRef]
- Dinh, T.P.; Carpenter, D.; Leslie, F.M.; Freund, T.F.; Katona, I.; Sensi, S.L.; Kathuria, S.; Piomelli, D. Brain monoglyceride lipase participating in endocannabinoid inactivation. Proc. Natl. Acad. Sci. USA 2002, 99, 10819–10824. [Google Scholar] [CrossRef]
- Basavarajappa, B.S. Endocannabinoid System and Alcohol Abuse Disorders. In Recent Advances in Cannabinoid Physiology and Pathology; Bukiya, A.N., Ed.; Nature Springer: Cham, Switzerland, 2019; 25p. [Google Scholar]
- Basavarajappa, B.S.; Joshi, V.; Shivakumar, M.; Subbanna, S. Distinct functions of endogenous cannabinoid system in alcohol abuse disorders. J. Cereb. Blood Flow Metab. 2019, 176, 3085–3109. [Google Scholar] [CrossRef] [PubMed]
- Busquets-García, A.; Bolaños, J.P.; Marsicano, G. Metabolic Messengers: Endocannabinoids. Nat. Metab. 2022, 4, 848–855. [Google Scholar] [CrossRef] [PubMed]
- Ledent, C.; Valverde, O.; Cossu, G.; Petitet, F.; Aubert, J.-F.; Beslot, F.; Böhme, G.A.; Imperato, A.; Pedrazzini, T.; Roques, B.P.; et al. Unresponsiveness to Cannabinoids and Reduced Addictive Effects of Opiates in CB 1 Receptor Knockout Mice. Science 1999, 283, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, A.; Zimmer, A.M.; Hohmann, A.G.; Herkenham, M.; Bonner, T.I. Increased mortality, hypoactivity, and hypoalgesia in cannabinoid CB1 receptor knockout mice. Proc. Natl. Acad. Sci. USA 1999, 96, 5780–5785. [Google Scholar] [CrossRef]
- Mackie, K. Cannabinoid receptors as therapeutic targets. Annu. Rev. Pharmacol. Toxicol. 2006, 46, 101–122. [Google Scholar] [CrossRef]
- Mechoulam, R. (Ed.) The pharmacohistory of Cannabis sativa. In Cannabinoids as Therapeutic Agents; CRC Press: Boca Raton, FL, USA, 1986. [Google Scholar]
- Herkenham, M.; Lynn, A.B.; Little, M.D.; Johnson, M.R.; Melvin, L.S.; de Costa, B.R.; Rice, K.C. Cannabinoid receptor localization in brain. Proc. Natl. Acad. Sci. USA 1990, 87, 1932–1936. [Google Scholar] [CrossRef] [PubMed]
- Tsou, K.; Brown, S.; Sañudo-Peña, M.; Mackie, K.; Walker, J. Immunohistochemical distribution of cannabinoid CB1 receptors in the rat central nervous system. Neuroscience 1998, 83, 393–411. [Google Scholar] [CrossRef]
- Schlicker, E.; Kathmann, M. Modulation of transmitter release via presynaptic cannabinoid receptors. Trends Pharmacol. Sci. 2001, 22, 565–572. [Google Scholar] [CrossRef]
- Wilson, R.I.; Nicoll, R.A. Endocannabinoid Signaling in the Brain. Science 2002, 296, 678–682. [Google Scholar] [CrossRef]
- Matias, I.; Di Marzo, V. Endocannabinoid synthesis and degradation, and their regulation in the framework of energy balance. J. Endocrinol. Investig. 2006, 29, 15–26. [Google Scholar]
- Lunn, C.A.; Reich, E.P.; Bober, L. Targeting the CB2 receptor for immune modulation. Expert Opin. Ther. Targets 2006, 10, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Onaivi, E.S.; Ishiguro, H.; Gong, J.P.; Patel, S.; Perchuk, A.; Meozzi, P.A.; Myers, L.; Mora, Z.; Tagliaferro, P.; Gardner, E.; et al. Discovery of the Presence and Functional Expression of Cannabinoid CB2 Receptors in Brain. Ann. N. Y. Acad. Sci. 2006, 1074, 514–536. [Google Scholar] [CrossRef]
- Gong, J.-P.; Onaivi, E.S.; Ishiguro, H.; Liu, Q.-R.; Tagliaferro, P.A.; Brusco, A.; Uhl, G.R. Cannabinoid CB2 receptors: Immunohistochemical localization in rat brain. Brain Res. 2006, 1071, 10–23. [Google Scholar] [CrossRef]
- Van Sickle, M.D.; Duncan, M.; Kingsley, P.J.; Mouihate, A.; Urbani, P.; Mackie, K.; Stella, N.; Makriyannis, A.; Piomelli, D.; Davison, J.S.; et al. Identification and Functional Characterization of Brainstem Cannabinoid CB 2 Receptors. Science 2005, 310, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Zavaleta, R.; Cortés, H.; Avalos-Fuentes, J.A.; García, U.; Vila, J.S.; Erlij, D.; Florán, B. Presynaptic cannabinoid CB2 receptors modulate [3 H]-Glutamate release at subthalamo-nigral terminals of the rat. Synapse 2018, 72, e22061. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hua, T.; Vemuri, K.; Ho, J.-H.; Wu, Y.; Wu, L.; Popov, P.; Benchama, O.; Zvonok, N.; Locke, K.; et al. Crystal Structure of the Human Cannabinoid Receptor CB2. Cell 2019, 176, 459–467.e13. [Google Scholar] [CrossRef] [PubMed]
- Micale, V.; Stepan, J.; Jurik, A.; Pamplona, F.A.; Marsch, R.; Drago, F.; Eder, M.; Wotjak, C.T. Extinction of avoidance behavior by safety learning depends on endocannabinoid signaling in the hippocampus. J. Psychiatr. Res. 2017, 90, 46–59. [Google Scholar] [CrossRef]
- Ruehle, S.; Wager-Miller, J.; Straiker, A.; Farnsworth, J.; Murphy, M.N.; Loch, S.; Monory, K.; Mackie, K.; Lutz, B. Discovery and characterization of two novel CB1 receptor splice variants with modified N-termini in mouse. J. Neurochem. 2017, 142, 521–533. [Google Scholar] [CrossRef]
- Hoehe, M.R.; Caenazzo, L.; Martinez, M.M.; Hsieh, W.T.; Modi, W.S.; Gershon, E.S.; Bonner, T.I. Genetic and physical mapping of the human cannabinoid receptor gene to chromosome 6q14-q15. New Biol. 1991, 3, 880–885. [Google Scholar]
- McCaw, E.A.; Hu, H.; Gomez, G.T.; Hebb, A.L.; Kelly, M.E.; Denovan-Wright, E.M. Structure, expression and regulation of the cannabinoid receptor gene (CB1) in Huntington’s disease transgenic mice. Eur. J. Biochem. 2004, 271, 4909–4920. [Google Scholar] [CrossRef]
- Zhang, P.-W.; Ishiguro, H.; Ohtsuki, T.; Hess, J.; Carillo, F.; Walther, D.; Onaivi, E.S.; Arinami, T.; Uhl, G.R. Human cannabinoid receptor 1: 5′ exons, candidate regulatory regions, polymorphisms, haplotypes and association with polysubstance abuse. Mol. Psychiatry 2004, 9, 916–931. [Google Scholar] [CrossRef] [PubMed]
- Miller, L.K.; Devi, L.A. The Highs and Lows of Cannabinoid Receptor Expression in Disease: Mechanisms and Their Therapeutic Implications. Pharmacol. Rev. 2011, 63, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Tsunoda, T.; Takagi, T. Estimating transcription factor bindability on DNA. Bioinformatics 1999, 15, 622–630. [Google Scholar] [CrossRef] [PubMed]
- Tao, R.; Li, C.; Jaffe, A.E.; Shin, J.H.; Deep-Soboslay, A.; Yamin, R.; Weinberger, D.R.; Hyde, T.M.; Kleinman, J.E. Cannabinoid receptor CNR1 expression and DNA methylation in human prefrontal cortex, hippocampus and caudate in brain development and schizophrenia. Transl. Psychiatry 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Nicoll, G.; Davidson, S.; Shanley, L.; Hing, B.; Lear, M.; McGuffin, P.; Ross, R.; MacKenzie, A. Allele-specific Differences in Activity of a Novel Cannabinoid Receptor 1 (CNR1) Gene Intronic Enhancer in Hypothalamus, Dorsal Root Ganglia, and Hippocampus. J. Biol. Chem. 2012, 287, 12828–12834. [Google Scholar] [CrossRef]
- Hay, E.A.; McEwan, A.; Wilson, D.; Barrett, P.; D’Agostino, G.; Pertwee, R.G.; MacKenzie, A. Disruption of an enhancer associated with addictive behaviour within the cannabinoid receptor-1 gene suggests a possible role in alcohol intake, cannabinoid response and anxiety-related behaviour. Psychoneuroendocrinology 2019, 109, 104407. [Google Scholar] [CrossRef]
- Katona, I.; Rancz, E.A.; Acsády, L.; Ledent, C.; Mackie, K.; Hajos, N.; Freund, T.F. Distribution of CB1 Cannabinoid Receptors in the Amygdala and their Role in the Control of GABAergic Transmission. J. Neurosci. 2001, 21, 9506–9518. [Google Scholar] [CrossRef]
- Stincic, T.L.; Hyson, R.L. Localization of CB1 cannabinoid receptor mRNA in the brain of the chick (Gallus domesticus). Brain Res. 2008, 1245, 61–73. [Google Scholar] [CrossRef]
- Van Waes, V.; Beverley, J.A.; Siman, H.; Tseng, K.Y.; Steiner, H. CB1 Cannabinoid Receptor Expression in the Striatum: Association with Corticostriatal Circuits and Developmental Regulation. Front. Pharmacol. 2012, 3, 21. [Google Scholar] [CrossRef]
- Jordan, B.; Devi, L.A. G-protein-coupled receptor heterodimerization modulates receptor function. Nature 1999, 399, 697–700. [Google Scholar] [CrossRef]
- Joshi, N.; Onaivi, E.S. Endocannabinoid System Components: Overview and Tissue Distribution. Adv. Exp. Med. Biol. 2019, 1162, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Munro, S.; Thomas, K.L.; Abu-Shaar, M. Molecular characterization of a peripheral receptor for cannabinoids. Nature 1993, 365, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Onaivi, E.S.; Leonard, C.M.; Ishiguro, H.; Zhang, P.W.; Lin, Z.; Akinshola, B.E.; Uhl, G.R. Endocannabinoids and cannabinoid receptor genetics. Prog. Neurobiol. 2002, 66, 307–344. [Google Scholar] [CrossRef]
- Shire, D.; Calandra, B.; Rinaldi-Carmona, M.; Oustric, D.; Pessegue, B.; Bonnin-Cabanne, O.; Le Fur, G.; Caput, D.; Ferrara, P. Molecular cloning, expression and function of the murine CB2 peripheral cannabinoid receptor. Biochim. Biophys. Acta (BBA) Gene Struct. Expr. 1996, 1307, 132–136. [Google Scholar] [CrossRef]
- Brown, S.M.; Wager-Miller, J.; Mackie, K. Cloning and molecular characterization of the rat CB2 cannabinoid receptor. Biochim. Biophys. Acta (BBA)-Gene Struct. Expr. 2002, 1576, 255–264. [Google Scholar] [CrossRef]
- Chen, D.-J.; Gao, M.; Gao, F.-F.; Su, Q.-X.; Wu, J. Brain cannabinoid receptor 2: Expression, function and modulation. Acta Pharmacol. Sin. 2017, 38, 312–316. [Google Scholar] [CrossRef]
- Ishiguro, H.; Kibret, B.G.; Horiuchi, Y.; Onaivi, E.S. Potential Role of Cannabinoid Type 2 Receptors in Neuropsychiatric and Neurodegenerative Disorders. Front. Psychiatry 2022, 13, 828895. [Google Scholar] [CrossRef]
- Jordan, C.J.; Xi, Z.-X. Progress in brain cannabinoid CB2 receptor research: From genes to behavior. Neurosci. Biobehav. Rev. 2019, 98, 208–220. [Google Scholar] [CrossRef]
- Li, Y.; Kim, J. Neuronal expression of CB2 cannabinoid receptor mRNAs in the mouse hippocampus. Neuroscience 2015, 311, 253–267. [Google Scholar] [CrossRef]
- Ghosh, K.; Zhang, G.-F.; Chen, H.; Chen, S.-R.; Pan, H.-L. Cannabinoid CB2 receptors are upregulated via bivalent histone modifications and control primary afferent input to the spinal cord in neuropathic pain. J. Biol. Chem. 2022, 298, 101999. [Google Scholar] [CrossRef]
- Liu, Q.R.; Pan, C.H.; Hishimoto, A.; Li, C.Y.; Xi, Z.X.; Llorente-Berzal, A.; Viveros, M.P.; Ishiguro, H.; Arinami, T.; Onaivi, E.S.; et al. Species differences in cannabinoid receptor 2 (CNR2 gene): Identification of novel human and rodent CB2 isoforms, differential tissue expression and regulation by cannabinoid receptor ligands. Genes Brain Behav. 2009, 8, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Canseco-Alba, A.; Sanabria, B.; Hammouda, M.; Bernadin, R.; Mina, M.; Liu, Q.-R.; Onaivi, E.S. Cell-Type Specific Deletion of CB2 Cannabinoid Receptors in Dopamine Neurons Induced Hyperactivity Phenotype: Possible Relevance to Attention-Deficit Hyperactivity Disorder. Front. Psychiatry 2022, 12, 803394. [Google Scholar] [CrossRef] [PubMed]
- Sherwood, T.A.; Nong, L.; Agudelo, M.; Newton, C.; Widen, R.; Klein, T.W. Identification of Transcription Start Sites and Preferential Expression of Select CB2 Transcripts in Mouse and Human B Lymphocytes. J. Neuroimmune Pharmacol. 2009, 4, 476–488. [Google Scholar] [CrossRef]
- Onaivi, E.S.; Ishiguro, H.; Gu, S.; Liu, Q.-R. CNS effects of CB2 cannabinoid receptors: Beyond neuro-immuno-cannabinoid activity. J. Psychopharmacol. 2011, 26, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Galán-Ganga, M.; del Río, R.; Jiménez-Moreno, N.; Díaz-Guerra, M.; Lastres-Becker, I. Cannabinoid CB2 Receptor Modulation by the Transcription Factor NRF2 is Specific in Microglial Cells. Cell. Mol. Neurobiol. 2019, 40, 167–177. [Google Scholar] [CrossRef]
- Gomes, T.M.; da Silva, D.D.; Carmo, H.; Carvalho, F.; Silva, J.P. Epigenetics and the endocannabinoid system signaling: An intricate interplay modulating neurodevelopment. Pharmacol. Res. 2020, 162, 105237. [Google Scholar] [CrossRef]
- Basavarajappa, B.S.; Subbanna, S. Epigenetic Mechanisms in Developmental Alcohol-Induced Neurobehavioral Deficits. Brain Sci. 2016, 6, 12. [Google Scholar] [CrossRef]
- Basavarajappa, B.S.; Subbanna, S. Potential Mechanisms Underlying the Deleterious Effects of Synthetic Cannabinoids Found in Spice/K2 Products. Brain Sci. 2019, 9, 14. [Google Scholar] [CrossRef]
- Kukreja, L.; Li, C.J.; Ezhilan, S.; Iyer, V.R.; Kuo, J.S. Emerging Epigenetic Therapies for Brain Tumors. Neuromol. Med. 2021, 24, 41–49. [Google Scholar] [CrossRef]
- Murshid, N.M.; Lubis, F.A.; Makpol, S. Epigenetic Changes and Its Intervention in Age-Related Neurodegenerative Diseases. Cell. Mol. Neurobiol. 2020, 42, 577–595. [Google Scholar] [CrossRef]
- Morselli, M.; Dieci, G. Epigenetic regulation of human non-coding RNA gene transcription. Biochem. Soc. Trans. 2022, 50, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.U.; Ma, D.K.; Mo, H.; Ball, M.; Jang, M.-H.; Bonaguidi, M.A.; Balazer, J.A.; Eaves, H.L.; Xie, B.; Ford, E.; et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat. Neurosci. 2011, 14, 1345–1351. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.U.; Su, Y.; Zhong, C.; Ming, G.-L.; Song, H. Hydroxylation of 5-Methylcytosine by TET1 Promotes Active DNA Demethylation in the Adult Brain. Cell 2011, 145, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Nabel, C.S.; Kohli, R.M. Demystifying DNA Demethylation. Science 2011, 333, 1229–1230. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.C.; Zhang, Y. Active DNA demethylation: Many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 2010, 11, 607–620. [Google Scholar] [CrossRef]
- Szulwach, K.E.; Li, X.; Li, Y.; Song, C.X.; Wu, H.; Dai, Q.; Irier, H.; Upadhyay, A.K.; Gearing, M.; Levey, A.I.; et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 2011, 14, 1607–1616. [Google Scholar] [CrossRef]
- Kubiura, M.; Okano, M.; Kimura, H.; Kawamura, F.; Tada, M. Chromosome-wide regulation of euchromatin-specific 5mC to 5hmC conversion in mouse ES cells and female human somatic cells. Chromosom. Res. 2012, 20, 837–848. [Google Scholar] [CrossRef]
- Nan, X.; Ng, H.H.; Johnson, C.A.; Laherty, C.D.; Turner, B.M.; Eisenman, R.N.; Bird, A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 1998, 393, 386–389. [Google Scholar] [CrossRef]
- Sarraf, S.A.; Stancheva, I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol. Cell. 2004, 15, 595–605. [Google Scholar] [CrossRef]
- Tao, J.; Hu, K.; Chang, Q.; Wu, H.; Sherman, N.E.; Martinowich, K.; Klose, R.J.; Schanen, C.; Jaenisch, R.; Wang, W.; et al. Phosphorylation of MeCP2 at Serine 80 regulates its chromatin association and neurological function. Proc. Natl. Acad. Sci. USA 2009, 106, 4882–4887. [Google Scholar] [CrossRef]
- Zhou, Z.; Hong, E.J.; Cohen, S.; Zhao, W.-N.; Ho, H.-Y.H.; Schmidt, L.; Chen, W.G.; Lin, Y.; Savner, E.; Griffith, E.C.; et al. Brain-Specific Phosphorylation of MeCP2 Regulates Activity-Dependent Bdnf Transcription, Dendritic Growth, and Spine Maturation. Neuron 2006, 52, 255–269. [Google Scholar] [CrossRef] [PubMed]
- Laprairie, R.; Kelly, M.; Denovan-Wright, E. The dynamic nature of type 1 cannabinoid receptor (CB1) gene transcription. J. Cereb. Blood Flow Metab. 2012, 167, 1583–1595. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wang, H.; Ning, W.; Backlund, M.G.; Dey, S.K.; DuBois, R.N. Loss of Cannabinoid Receptor 1 Accelerates Intestinal Tumor Growth. Cancer Res. 2008, 68, 6468–6476. [Google Scholar] [CrossRef] [PubMed]
- Xia, D.; Wang, D.; Kim, S.-H.; Katoh, H.; Dubois, R.N. Prostaglandin E2 promotes intestinal tumor growth via DNA methylation. Nat. Med. 2012, 18, 224–226. [Google Scholar] [CrossRef]
- Franklin, T.B.; Russig, H.; Weiss, I.C.; Gräff, J.; Linder, N.; Michalon, A.; Vizi, S.; Mansuy, I.M. Epigenetic Transmission of the Impact of Early Stress Across Generations. Biol. Psychiatry 2010, 68, 408–415. [Google Scholar] [CrossRef]
- Viveros, M.-P.; Marco, E.-M.; Llorente, R.; López-Gallardo, M. Endocannabinoid System and Synaptic Plasticity: Implications for Emotional Responses. Neural Plast. 2007, 2007, 1–12. [Google Scholar] [CrossRef]
- Börner, C.; Martella, E.; Höllt, V.; Kraus, J. Regulation of Opioid and Cannabinoid Receptor Genes in Human Neuroblastoma and T Cells by the Epigenetic Modifiers Trichostatin A and 5-Aza-2′-Deoxycytidine. Neuroimmunomodulation 2012, 19, 180–186. [Google Scholar] [CrossRef]
- Rotter, A.; Bayerlein, K.; Hansbauer, M.; Weiland, J.; Sperling, W.; Kornhuber, J.; Biermann, T. CB1 and CB2 Receptor Expression and Promoter Methylation in Patients with Cannabis Dependence. Eur. Addict. Res. 2012, 19, 13–20. [Google Scholar] [CrossRef]
- Di Francesco, A.; Falconi, A.; Di Germanio, C.; Di Bonaventura, M.V.M.; Costa, A.; Caramuta, S.; Del Carlo, M.; Compagnone, D.; Dainese, E.; Cifani, C.; et al. Extravirgin olive oil up-regulates CB1 tumor suppressor gene in human colon cancer cells and in rat colon via epigenetic mechanisms. J. Nutr. Biochem. 2015, 26, 250–258. [Google Scholar] [CrossRef]
- Hong, S.; Zheng, G.; Wiley, J.W. Epigenetic Regulation of Genes That Modulate Chronic Stress-Induced Visceral Pain in the Peripheral Nervous System. Gastroenterology 2015, 148, 148–157.e7. [Google Scholar] [CrossRef]
- D’Addario, C.; Micale, V.; Di Bartolomeo, M.; Stark, T.; Pucci, M.; Sulcova, A.; Palazzo, M.; Babinska, Z.; Cremaschi, L.; Drago, F.; et al. A preliminary study of endocannabinoid system regulation in psychosis: Distinct alterations of CNR1 promoter DNA methylation in patients with schizophrenia. Schizophr. Res. 2017, 188, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Pucci, M.; Di Bonaventura, M.V.M.; Vezzoli, V.; Zaplatic, E.; Massimini, M.; Mai, S.; Sartorio, A.; Scacchi, M.; Persani, L.; Maccarrone, M.; et al. Preclinical and Clinical Evidence for a Distinct Regulation of Mu Opioid and Type 1 Cannabinoid Receptor Genes Expression in Obesity. Front. Genet. 2019, 10, 523. [Google Scholar] [CrossRef] [PubMed]
- D’Addario, C.; Ms, E.Z.; Giunti, E.; Pucci, M.; Di Bonaventura, M.V.M.; Scherma, M.; Dainese, E.; Maccarrone, M.; Nilsson, I.A.; Cifani, C.; et al. Epigenetic regulation of the cannabinoid receptor CB1 in an activity-based rat model of anorexia nervosa. Int. J. Eat. Disord. 2020, 53, 702–716. [Google Scholar] [CrossRef] [PubMed]
- Mancino, S.; Burokas, A.; Gutiérrez-Cuesta, J.; Gutiérrez-Martos, M.; Martín-García, E.; Pucci, M.; Falconi, A.; D’Addario, C.; Maccarrone, M.; Maldonado, R. Epigenetic and Proteomic Expression Changes Promoted by Eating Addictive-Like Behavior. Neuropsychopharmacology 2015, 40, 2788–2800. [Google Scholar] [CrossRef]
- Subbanna, S.; Shivakumar, M.; Psychoyos, D.; Xie, S.; Basavarajappa, B.S. Anandamide-CB1 Receptor Signaling Contributes to Postnatal Ethanol-Induced Neonatal Neurodegeneration, Adult Synaptic, and Memory Deficits. J. Neurosci. 2013, 33, 6350–6366. [Google Scholar] [CrossRef]
- Nagre, N.N.; Subbanna, S.; Shivakumar, M.; Psychoyos, D.; Basavarajappa, B.S. CB1-receptor knockout neonatal mice are protected against ethanol-induced impairments of DNMT1, DNMT3A, and DNA methylation. J. Neurochem. 2015, 132, 429–442. [Google Scholar] [CrossRef]
- Smith, R.C.; Sershen, H.; Janowsky, D.S.; Lajtha, A.; Grieco, M.; Gangoiti, J.A.; Gertsman, I.; Johnson, W.S.; Marcotte, T.D.; Davis, J.M. Changes in Expression of DNA-Methyltransferase and Cannabinoid Receptor mRNAs in Blood Lymphocytes After Acute Cannabis Smoking. Front. Psychiatry 2022, 13, 887700. [Google Scholar] [CrossRef]
- Innocenzi, E.; De Domenico, E.; Ciccarone, F.; Zampieri, M.; Rossi, G.; Cicconi, R.; Bernardini, R.; Mattei, M.; Grimaldi, P. Paternal activation of CB2 cannabinoid receptor impairs placental and embryonic growth via an epigenetic mechanism. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Parastouei, K.; Aarabi, M.H.; Hamidi, G.A.; Nasehi, Z.; Arani, S.K.; Jozi, F.; Shahabodin, M.E. A CB2 Receptor Agonist Reduces the Production of Inflammatory Mediators and Improves Locomotor Activity in Experimental Autoimmune Encephalomyelitis. Rep. Biochem. Mol. Biol. 2022, 11, 1–9. [Google Scholar] [CrossRef]
- Strisciuglio, C.; Creoli, M.; Tortora, C.; Martinelli, M.; Miele, E.; Paino, S.; Luongo, L.; Rossi, F. Increased expression of CB2 receptor in the intestinal biopsies of children with inflammatory bowel disease. Pediatr. Res. 2022, 1–6. [Google Scholar] [CrossRef]
- Kiran, S.; Rakib, A.; Moore, B.M.; Singh, U.P. Cannabinoid Receptor 2 (CB2) Inverse Agonist SMM-189 Induces Expression of Endogenous CB2 and Protein Kinase A That Differentially Modulates the Immune Response and Suppresses Experimental Colitis. Pharmaceutics 2022, 14, 936. [Google Scholar] [CrossRef]
- Ten-Blanco, M.; Flores, Á.; Pereda-Pérez, I.; Piscitelli, F.; Izquierdo-Luengo, C.; Cristino, L.; Romero, J.; Hillard, C.J.; Maldonado, R.; Di Marzo, V.; et al. Amygdalar CB2 cannabinoid receptor mediates fear extinction deficits promoted by orexin-A/hypocretin-1. Biomed. Pharmacother. 2022, 149, 112925. [Google Scholar] [CrossRef]
- Jayanthi, S.; Peesapati, R.; McCoy, M.T.; Ladenheim, B.; Cadet, J.L. Footshock-Induced Abstinence from Compulsive Methamphetamine Self-administration in Rat Model Is Accompanied by Increased Hippocampal Expression of Cannabinoid Receptors (CB1 and CB2). Mol. Neurobiol. 2022, 59, 1238–1248. [Google Scholar] [CrossRef]
- Paradisi, A.; Pasquariello, N.; Barcaroli, D.; Maccarrone, M. Anandamide Regulates Keratinocyte Differentiation by Inducing DNA Methylation in a CB1 Receptor-dependent Manner. J. Biol. Chem. 2008, 283, 6005–6012. [Google Scholar] [CrossRef] [PubMed]
- Molina, P.E.; Amedee, A.; LeCapitaine, N.J.; Zabaleta, J.; Mohan, M.; Winsauer, P.; Stouwe, C.V. Cannabinoid Neuroimmune Modulation of SIV Disease. J. Neuroimmune Pharmacol. 2011, 6, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Gobira, P.H.; Roncalho, A.L.; Silva, N.R.; Silote, G.P.; Sales, A.J.; Joca, S.R. Adolescent cannabinoid exposure modulates the vulnerability to cocaine-induced conditioned place preference and DNMT3a expression in the prefrontal cortex in Swiss mice. Psychopharmacology 2021, 238, 3107–3118. [Google Scholar] [CrossRef] [PubMed]
- Schrott, R.; Rajavel, M.; Acharya, K.; Huang, Z.; Acharya, C.; Hawkey, A.; Pippen, E.; Lyerly, H.K.; Levin, E.D.; Murphy, S.K. Sperm DNA methylation altered by THC and nicotine: Vulnerability of neurodevelopmental genes with bivalent chromatin. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Jahn, K.; Heese, A.; Kebir, O.; Groh, A.; Bleich, S.; Krebs, M.O.; Frieling, H. Differential Methylation Pattern of Schizophrenia Candidate Genes in Tetrahydrocannabinol-Consuming Treatment-Resistant Schizophrenic Patients Compared to Non-Consumer Patients and Healthy Controls. Neuropsychobiology 2020, 80, 36–44. [Google Scholar] [CrossRef]
- Tomas-Roig, J.; Benito, E.; Agis-Balboa, R.; Piscitelli, F.; Hoyer-Fender, S.; Di Marzo, V.; Havemann-Reinecke, U. Chronic exposure to cannabinoids during adolescence causes long-lasting behavioral deficits in adult mice. Addict. Biol. 2016, 22, 1778–1789. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Wu, H.; Feng, X.; Chen, Y.; Lv, Z.; Kota, V.G.; Chen, J.; Wu, W.; Lu, Y.; Liu, H.; et al. DNA Methylation of Cannabinoid Receptor Interacting Protein 1 Promotes Pathogenesis of Intrahepatic Cholangiocarcinoma Through Suppressing Parkin-Dependent Pyruvate Kinase M2 Ubiquitination. Hepatology 2020, 73, 1816–1835. [Google Scholar] [CrossRef]
- Subbanna, S.; Nagre, N.N.; Shivakumar, M.; Joshi, V.; Psychoyos, D.; Kutlar, A.; Umapathy, N.S.; Basavarajappa, B.S. CB1R-Mediated Activation of Caspase-3 Causes Epigenetic and Neurobehavioral Abnormalities in Postnatal Ethanol-Exposed Mice. Front. Mol. Neurosci. 2018, 11, 45. [Google Scholar] [CrossRef] [PubMed]
- Pedrazzi, J.F.; Sales, A.J.; Guimarães, F.S.; Joca, S.R.; Crippa, J.A.; Del Bel, E. Cannabidiol prevents disruptions in sensorimotor gating induced by psychotomimetic drugs that last for 24-h with probable involvement of epigenetic changes in the ventral striatum. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2021, 111, 110352. [Google Scholar] [CrossRef] [PubMed]
- Wanner, N.M.; Colwell, M.; Drown, C.; Faulk, C. Subacute cannabidiol alters genome-wide DNA methylation in adult mouse hippocampus. Environ. Mol. Mutagen. 2020, 61, 890–900. [Google Scholar] [CrossRef]
- Elliott, E.; Manashirov, S.; Zwang, R.; Gil, S.; Tsoory, M.; Shemesh, Y.; Chen, A. Dnmt3a in the Medial Prefrontal Cortex Regulates Anxiety-Like Behavior in Adult Mice. J. Neurosci. 2016, 36, 730–740. [Google Scholar] [CrossRef]
- Henikoff, S.; Smith, M.M. Histone Variants and Epigenetics. Cold Spring Harb. Perspect. Biol. 2015, 7, a019364. [Google Scholar] [CrossRef]
- Park, J.; Lee, K.; Kim, K.; Yi, S.-J. The role of histone modifications: From neurodevelopment to neurodiseases. Signal Transduct. Target. Ther. 2022, 7, 1–23. [Google Scholar] [CrossRef]
- Kornberg, R.D.; Lorch, Y. Twenty-Five Years of the Nucleosome, Fundamental Particle of the Eukaryote Chromosome. Cell 1999, 98, 285–294. [Google Scholar] [CrossRef]
- Lorch, Y.; Zhang, M.; Kornberg, R.D. Histone Octamer Transfer by a Chromatin-Remodeling Complex. Cell 1999, 96, 389–392. [Google Scholar] [CrossRef]
- Goll, M.G.; Bestor, T.H. Histone modification and replacement in chromatin activation: Figure 1. Genes Dev. 2002, 16, 1739–1742. [Google Scholar] [CrossRef]
- Grant, P.A. A tale of histone modifications. Genome Biol. 2001, 2, 1–6. [Google Scholar] [CrossRef]
- Smolle, M.; Workman, J.L. Transcription-associated histone modifications and cryptic transcription. Biochim. Biophys. Acta 2012, 1829, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Joseph, F.M.; Young, N.L. Histone variant-specific post-translational modifications. Semin. Cell Dev. Biol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Lomazzo, E.; König, F.; Abassi, L.; Jelinek, R.; Lutz, B. Chronic stress leads to epigenetic dysregulation in the neuropeptide-Y and cannabinoid CB1 receptor genes in the mouse cingulate cortex. Neuropharmacology 2017, 113, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Subbanna, S.; Nagre, N.N.; Umapathy, N.S.; Pace, B.; Basavarajappa, B.S. Ethanol Exposure Induces Neonatal Neurodegeneration by Enhancing CB1R Exon1 Histone H4K8 Acetylation and Up-regulating CB1R Function causing Neurobehavioral Abnormalities in Adult Mice. Int. J. Neuropsychopharmacol. 2014, 18, pyu028. [Google Scholar] [CrossRef] [PubMed]
- Shivakumar, M.; Subbanna, S.; Joshi, V.; Basavarajappa, B.S. Postnatal Ethanol Exposure Activates HDAC-Mediated Histone Deacetylation, Impairs Synaptic Plasticity Gene Expression and Behavior in Mice. Int. J. Neuropsychopharmacol. 2020, 23, 324–338. [Google Scholar] [CrossRef]
- Luo, Y.; Zhang, J.; Chen, L.; Chen, S.-R.; Chen, H.; Zhang, G.; Pan, H.-L. Histone methyltransferase G9a diminishes expression of cannabinoid CB1 receptors in primary sensory neurons in neuropathic pain. J. Biol. Chem. 2020, 295, 3553–3562. [Google Scholar] [CrossRef]
- Nogueira, D.D.S.; Bourdy, R.; Alcala-Vida, R.; Filliol, D.; Andry, V.; Goumon, Y.; Zwiller, J.; Romieu, P.; Merienne, K.; Olmstead, M.C.; et al. Hippocampal Cannabinoid 1 Receptors Are Modulated Following Cocaine Self-administration in Male Rats. Mol. Neurobiol. 2022, 59, 1896–1911. [Google Scholar] [CrossRef]
- De Sa Nogueira, D.; Merienne, K.; Befort, K. Neuroepigenetics and addictive behaviors: Where do we stand? Neurosci. Biobehav. Rev. 2019, 106, 58–72. [Google Scholar] [CrossRef]
- Renthal, W.; Nestler, E.J. Epigenetic mechanisms in drug addiction. Trends Mol. Med. 2008, 14, 341–350. [Google Scholar] [CrossRef]
- Dhopeshwarkar, A.; Mackie, K. CB2 Cannabinoid receptors as a therapeutic target-what does the future hold? Mol. Pharmacol. 2014, 86, 430–437. [Google Scholar] [CrossRef]
- Tang, Y.; Wolk, B.; Britch, S.C.; Craft, R.M.; Kendall, D.A. Anti-inflammatory and antinociceptive effects of the selective cannabinoid CB2 receptor agonist ABK5. J. Pharmacol. Sci. 2020, 145, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; VHegde, L.; Rao, R.; Zhang, J.; Nagarkatti, P.S.; Nagarkatti, M. Histone modifications are associated with Delta9-tetrahydrocannabinol-mediated alterations in antigen-specific T cell responses. J. Biol. Chem. 2014, 289, 18707–18718. [Google Scholar] [CrossRef] [PubMed]
- Prini, P.; Rusconi, F.; Zamberletti, E.; Gabaglio, M.; Penna, F.; Fasano, M.; Battaglioli, E.; Parolaro, D.; Rubino, T. Adolescent THC exposure in female rats leads to cognitive deficits through a mechanism involving chromatin modifications in the prefrontal cortex. J. Psychiatry Neurosci. 2018, 43, 87–101. [Google Scholar] [CrossRef] [PubMed]
- Bilkei-Gorzo, A.; Albayram, O.; Draffehn, A.; Michel, K.; Piyanova, A.; Oppenheimer, H.; Dvir-Ginzberg, M.; Rácz, I.; Ulas, T.; Imbeault, S.; et al. A chronic low dose of Delta(9)-tetrahydrocannabinol (THC) restores cognitive function in old mice. Nat. Med. 2017, 23, 782–787. [Google Scholar] [CrossRef]
- Yang, X.; Bam, M.; Nagarkatti, P.S.; Nagarkatti, M. Cannabidiol Regulates Gene Expression in Encephalitogenic T cells Using Histone Methylation and noncoding RNA during Experimental Autoimmune Encephalomyelitis. Sci. Rep. 2019, 9, 15780. [Google Scholar] [CrossRef]
- Todd, S.M.; Zhou, C.; Clarke, D.J.; Chohan, T.W.; Bahceci, D.; Arnold, J.C. Interactions between cannabidiol and Delta(9)-THC following acute and repeated dosing: Rebound hyperactivity, sensorimotor gating and epigenetic and neuroadaptive changes in the mesolimbic pathway. Eur. Neuropsychopharmacol. 2017, 27, 132–145. [Google Scholar] [CrossRef]
- Pastrana-Trejo, J.C.; Duarte-Aké, F.; Us-Camas, R.; De-La-Peña, C.; Parker, L.; Pertwee, R.G.; Murillo-Rodríguez, E. Effects on the Post-translational Modification of H3K4Me3, H3K9ac, H3K9Me2, H3K27Me3, and H3K36Me2 Levels in Cerebral Cortex, Hypothalamus and Pons of Rats after a Systemic Administration of Cannabidiol: A Preliminary Study. Central Nerv. Syst. Agents Med. Chem. 2021, 21, 142–147. [Google Scholar] [CrossRef]
- Hunter, R.G.; McCarthy, K.J.; Milne, T.A.; Pfaff, D.W.; McEwen, B.S. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc. Natl. Acad. Sci. USA 2009, 106, 20912–20917. [Google Scholar] [CrossRef]
- Tran, L.; Schulkin, J.; Ligon, C.O.; Meerveld, B.G.-V. Epigenetic modulation of chronic anxiety and pain by histone deacetylation. Mol. Psychiatry 2014, 20, 1219–1231. [Google Scholar] [CrossRef]
- Weaver, I.C.G.; Cervoni, N.; Champagne, F.A.; D’Alessio, A.C.; Sharma, S.; Seckl, J.R.; Dymov, S.; Szyf, M.; Meaney, M.J. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004, 7, 847–854. [Google Scholar] [CrossRef]
- Prini, P.; Penna, F.; Sciuccati, E.; Alberio, T.; Rubino, T. Chronic Delta(8)-THC Exposure Differently Affects Histone Modifications in the Adolescent and Adult Rat Brain. Int. J. Mol. Sci. 2017, 18, 2094. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Vasudevan, S. Posttranscriptional Upregulation by MicroRNAs. Wiley Interdiscip. Rev. RNA 2011, 3, 311–330. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Martinez, N.J.; Gregory, R.I. MicroRNA Gene Regulatory Pathways in the Establishment and Maintenance of ESC Identity. Cell Stem Cell 2010, 7, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef]
- Huang, W. MicroRNAs: Biomarkers, Diagnostics, and Therapeutics. Methods Mol. Biol. 2017, 1617, 57–67. [Google Scholar]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell. Physiol. 2015, 231, 25–30. [Google Scholar] [CrossRef]
- Paul, P.; Chakraborty, A.; Sarkar, D.; Langthasa, M.; Rahman, M.; Bari, M.; Singha, R.S.; Malakar, A.K.; Chakraborty, S. Interplay between miRNAs and human diseases. J. Cell. Physiol. 2017, 233, 2007–2018. [Google Scholar] [CrossRef]
- Tüfekci, K.U.; Öner, M.G.; Meuwissen, R.L.J.; Genç, Ş. The Role of MicroRNAs in Human Diseases. In miRNomics: MicroRNA Biology and Computational Analysis; Humana Press: Totowa, NJ, USA, 2013; pp. 33–50. [Google Scholar] [CrossRef]
- Roy, B.; Lee, E.; Li, T.; Rampersaud, M. Role of miRNAs in Neurodegeneration: From Disease Cause to Tools of Biomarker Discovery and Therapeutics. Genes 2022, 13, 425. [Google Scholar] [CrossRef]
- Leitão, A.L.; Enguita, F.J. A Structural View of miRNA Biogenesis and Function. Non-Coding RNA 2022, 8, 10. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Han, J.; Jung, Y. Pathological Contribution of Extracellular Vesicles and Their MicroRNAs to Progression of Chronic Liver Disease. Biology 2022, 11, 637. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Zhang, Q.; Jiang, L. Current Knowledge on Exosome Biogenesis, Cargo-Sorting Mechanism and Therapeutic Implications. Membranes 2022, 12, 498. [Google Scholar] [CrossRef] [PubMed]
- Kuwabara, T.; Hsieh, J.; Nakashima, K.; Taira, K.; Gage, F.H. A Small Modulatory dsRNA Specifies the Fate of Adult Neural Stem Cells. Cell 2004, 116, 779–793. [Google Scholar] [CrossRef]
- Poole, R.J.; Hobert, O. Early Embryonic Programming of Neuronal Left/Right Asymmetry in C. elegans. Curr. Biol. 2006, 16, 2279–2292. [Google Scholar] [CrossRef]
- Ronshaugen, M.; Biemar, F.; Piel, J.; Levine, M.; Lai, E.C. The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes Dev. 2005, 19, 2947–2952. [Google Scholar] [CrossRef]
- Schratt, G.M.; Tuebing, F.; Nigh, E.A.; Kane, C.G.; Sabatini, M.E.; Kiebler, M.; Greenberg, M.E. A brain-specific microRNA regulates dendritic spine development. Nature 2006, 439, 283–289. [Google Scholar] [CrossRef]
- Olde Loohuis, N.F.; Kos, A.; Martens, G.J.; Van Bokhoven, H.; Nadif Kasri, N.; Aschrafi, A. MicroRNA networks direct neuronal development and plasticity. Cell. Mol. Life Sci. 2012, 69, 89–102. [Google Scholar] [CrossRef]
- Davis, T.H.; Cuellar, T.L.; Koch, S.M.; Barker, A.J.; Harfe, B.D.; McManus, M.T.; Ullian, E.M. Conditional Loss of Dicer Disrupts Cellular and Tissue Morphogenesis in the Cortex and Hippocampus. J. Neurosci. 2008, 28, 4322–4330. [Google Scholar] [CrossRef]
- Stark, K.L.; Xu, B.; Bagchi, A.; Lai, W.-S.; Liu, H.; Hsu, R.; Wan, X.; Pavlidis, P.; Mills, A.A.; Karayiorgou, M.; et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat. Genet. 2008, 40, 751–760. [Google Scholar] [CrossRef]
- Li, M.; Qian, X.; Zhu, M.; Li, A.; Fang, M.; Zhu, Y.; Zhang, J. miR1273g3p promotes proliferation, migration and invasion of LoVo cells via cannabinoid receptor 1 through activation of ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Mol. Med. Rep. 2018, 17, 4619–4626. [Google Scholar] [PubMed]
- Sredni, S.T.; Huang, C.-C.; Suzuki, M.; Pundy, T.; Chou, P.; Tomita, T. Spontaneous involution of pediatric low-grade gliomas: High expression of cannabinoid receptor 1 (CNR1) at the time of diagnosis may indicate involvement of the endocannabinoid system. Child’s Nerv. Syst. 2016, 32, 2061–2067. [Google Scholar] [CrossRef] [PubMed]
- Möhnle, P.; Schütz, S.V.; Schmidt, M.; Hinske, C.; Hübner, M.; Heyn, J.; Beiras-Fernandez, A.; Kreth, S. MicroRNA-665 is involved in the regulation of the expression of the cardioprotective cannabinoid receptor CB2 in patients with severe heart failure. Biochem. Biophys. Res. Commun. 2014, 451, 516–521. [Google Scholar] [CrossRef] [PubMed]
- Chiarlone, A.; Börner, C.; Martín-Gómez, L.; Jiménez-González, A.; García-Concejo, A.; García-Bermejo, M.L.; Lorente, M.; Blázquez, C.; García-Taboada, E.; de Haro, A.; et al. MicroRNA let-7d is a target of cannabinoid CB 1 receptor and controls cannabinoid signaling. Neuropharmacology 2016, 108, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Bai, Z.; Yi, W.; Hu, Z.; Hao, J. Overexpression of miR-338-5p in exosomes derived from mesenchymal stromal cells provides neuroprotective effects by the Cnr1/Rap1/Akt pathway after spinal cord injury in rats. Neurosci. Lett. 2021, 761, 136124. [Google Scholar] [CrossRef]
- Miranda, K.; Mehrpouya-Bahrami, P.; Nagarkatti, P.S.; Nagarkatti, M. Cannabinoid Receptor 1 Blockade Attenuates Obesity and Adipose Tissue Type 1 Inflammation Through miR-30e-5p Regulation of Delta-Like-4 in Macrophages and Consequently Downregulation of Th1 Cells. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Li, L.; Xu, Y.; Zhao, M.; Gao, Z. Neuro-protective roles of long non-coding RNA MALAT1 in Alzheimer’s disease with the involvement of the microRNA-30b/CNR1 network and the following PI3K/AKT activation. Exp. Mol. Pathol. 2020, 117, 104545. [Google Scholar] [CrossRef]
- Xu, A.; Yang, Y.; Shao, Y.; Wu, M.; Sun, Y. Inhibiting effect of microRNA-187-3p on osteogenic differentiation of osteoblast precursor cells by suppressing cannabinoid receptor type 2. Differentiation 2019, 109, 9–15. [Google Scholar] [CrossRef]
- He, X.; Yang, L.; Huang, R.; Lin, L.; Shen, Y.; Cheng, L.; Jin, L.; Wang, S.; Zhu, R. Activation of CB2R with AM1241 ameliorates neurodegeneration via the Xist/miR-133b-3p/Pitx3 axis. J. Cell. Physiol. 2020, 235, 6032–6042. [Google Scholar] [CrossRef]
- Hegde, V.L.; Tomar, S.; Jackson, A.; Rao, R.; Yang, X.; Singh, U.P.; Singh, N.P.; Nagarkatti, P.S.; Nagarkatti, M. Distinct microRNA expression profile and targeted biological pathways in functional myeloid-derived suppressor cells induced by Delta9-tetrahydrocannabinol in vivo: Regulation of CCAAT/enhancer-binding protein alpha by microRNA-690. J. Biol. Chem. 2013, 288, 36810–36826. [Google Scholar] [CrossRef]
- Jackson, A.R.; Nagarkatti, P.; Nagarkatti, M. Anandamide Attenuates Th-17 Cell-Mediated Delayed-Type Hypersensitivity Response by Triggering IL-10 Production and Consequent microRNA Induction. PLoS ONE 2014, 9, e93954. [Google Scholar] [CrossRef] [PubMed]
- Simon, L.; Song, K.; Stouwe, C.V.; Hollenbach, A.; Amedee, A.; Mohan, M.; Winsauer, P.; Molina, P. Delta9-Tetrahydrocannabinol (Delta9-THC) Promotes Neuroimmune-Modulatory MicroRNA Profile in Striatum of Simian Immunodeficiency Virus (SIV)-Infected Macaques. J. Neuroimmune Pharmacol. 2016, 11, 192–213. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.; Nagarkatti, P.S.; Nagarkatti, M. Delta(9) Tetrahydrocannabinol attenuates Staphylococcal enterotoxin B-induced inflammatory lung injury and prevents mortality in mice by modulation of miR-17-92 cluster and induction of T-regulatory cells. Br. J. Pharmacol. 2015, 172, 1792–1806. [Google Scholar] [CrossRef] [PubMed]
- Chandra, L.C.; Kumar, V.; Torben, W.; Stouwe, C.V.; Winsauer, P.; Amedee, A.; Molina, P.E.; Mohan, M. Chronic administration of Delta9-tetrahydrocannabinol induces intestinal anti-inflammatory microRNA expression during acute simian immunodeficiency virus infection of rhesus macaques. J. Virol. 2015, 89, 1168–1181. [Google Scholar] [CrossRef] [PubMed]
- Al-Ghezi, Z.Z.; Miranda, K.; Nagarkatti, M.; Nagarkatti, P.S. Combination of Cannabinoids, Delta9- Tetrahydrocannabinol and Cannabidiol, Ameliorates Experimental Multiple Sclerosis by Suppressing Neuroinflammation Through Regulation of miRNA-Mediated Signaling Pathways. Front. Immunol. 2019, 10, 1921. [Google Scholar] [CrossRef]
- Martínez-Peña, A.A.; Lee, K.; Pereira, M.; Ayyash, A.; Petrik, J.J.; Hardy, D.B.; Holloway, A.C. Prenatal Exposure to Delta-9-tetrahydrocannabinol (THC) Alters the Expression of miR-122-5p and Its Target Igf1r in the Adult Rat Ovary. Int. J. Mol. Sci. 2022, 23, 8000. [Google Scholar] [CrossRef] [PubMed]
| Model | Treatment/Exposure | DNA Methylation at Cnr1 Promoter | Cnr1 Gene Expression | Reference |
|---|---|---|---|---|
| Colon cancer specimens | - | ↑ | ↓ | [75] |
| Human epithelial colon cell line LS-174T | Prostaglandin E2 | ↑ | ↓ | [76] |
| Rodents (PD 1 to 14) | Maternal separation | ↑ | - | [77] |
| Jurkat cells | 5-Aza-dC | ↓ | - | [79] |
| Humans | THC | ↑ | ↓ | [80] |
| Human colon cancer cells and rats | Extra-virgin olive oil (EVOO). Phenolic extracts (OPE) Hydroxytyrosol (HT) | ↓ | ↑ | [81] |
| Rats L6-S2 (DRG) | Chronic stress | ↑ | ↑ | [82] |
| Mice (PD 7) | Alcohol | ↓ | ↓ | [87] [88] |
| Schizophrenic patients | - | ↓ | ↑ | [83] |
| Rat (PFC) | Methylazoxymethanol acetate exposure | ↓ | ↑ | [83] |
| Human blood mononuclear cells from younger (<30 years old) human obese subjects | THC/alcohol | - | ↑ | [84] |
| Rat model | Anorexia | ↑ | ↓ | [85] |
| Schizophrenia patients | - | ↑ | ↓ | [36] |
| Model | Treatment | Histone Modifications | Gene Promoter | Reference |
|---|---|---|---|---|
| Mice (Cingulate cortex) | Chronic unpredictable stress | -Increased HDAC-2 -Decreased levels H3K9ac | Cnr1 | [115] |
| PD-7 Mice | Alcohol | -Increased H4K8ac -Decreased H3K9me2 | Cnr1 | [116] |
| PD-7 Mice | Alcohol | -Increased HDAC-1, HDAC-2, and HDAC-3 | Egr1, Arc | [117] |
| Rats | neuropathic pain | -Increased H3K9me2 | Cnr1 | [118] |
| Male Rats | Cocaine self-administration | -Increased H3K4me3, H3K27ac | Cnr1 | [119,125] |
| Female adolescent rats | THC | -Increased H3K9me3 -Increased Suv39H1, histone lysine methyltransferase, | ND | [125] |
| Female adolescent rats | THC | -Increased H3K14ac and H3K9me2 | ND | [125,133] |
| Mice | THC | Increased global H3K9ac, H4K12ac and reduced H3K9me3 | Klotho and Bdnf | [126] |
| THC and HAT inhibitor | ND | Decreased H3K9ac, synapsin 1, Klotho, and Bdnf expression | ||
| CD4+ T cells | CBD | Increased H3K4me3 and reduced H3K27me3 | IL-4, IL-5, and IL-13 | [127] |
| Adult male mice | CBD and THC | Increased H3K9ac and H3K14ac levels | ND | [128] |
| Adult rats Cerebral cortex Hypothalamus Pons | CBD | -Increased H3K4me3, H3K9ac, and H3K27me3 levels -Decreased H3K9ac levels -Decreased H3K4me3 levels | ND | [129] |
| Rats | Acute stress | -Increased H3K9me3 -Decreased H3K27me3 and H3K9me1 levels | ND | [130] |
| Chronic stress | -Increased H3K9me3 and decreased H3K27me3 and H3K4me3 levels | |||
| CBD | -Increased H3K9ac levels | |||
| Prolonged exposure to corticosteroids | Chronic anxiety and pain | -H3K9ac levels in the central amygdala | ND | [131] |
| Rats | Maternal care-induced anxiety-related behavior | -Decreased H3K9ac levels | glucocorticoid receptor gene (Nr3c1) | [132] |
| miRNA | Experimental Model | Target/Gene Expression | Reference |
|---|---|---|---|
| miRNA-1273g-3p | Human colon cancer cells | CB1 | [154] |
| hsa-miRNA-29b-3 | Low-grade glioma | [155] | |
| miR-494 | Myocardial cells | [156] | |
| miRNA let-7d | SH-SY5Y neuroblastoma cells Zebrafish, mice (cortex, striatum, and hippocampus) primary striatal neurons | [157] | |
| miR-30e-5p | Mice (obese model) | [159] | |
| miR-338-5p | Rats (spinal cord injury) | Cnr1 | [158] |
| MiRNA-30b | Rat models/cell models | [160] | |
| MiR-187-3p | Human osteoblastic precursor cells | Cnr2 | [161] |
| miR-133b-3p | PD mouse model | Xist and Pitx3 | [162] |
| miR-665 | Human (myocardial cells) | CB2 and Cnr2 | [156] |
| miRNA-690 | Mouse (MDSCs) | CB1 and CB2 | [163] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Basavarajappa, B.S.; Subbanna, S. Molecular Insights into Epigenetics and Cannabinoid Receptors. Biomolecules 2022, 12, 1560. https://doi.org/10.3390/biom12111560
Basavarajappa BS, Subbanna S. Molecular Insights into Epigenetics and Cannabinoid Receptors. Biomolecules. 2022; 12(11):1560. https://doi.org/10.3390/biom12111560
Chicago/Turabian StyleBasavarajappa, Balapal S., and Shivakumar Subbanna. 2022. "Molecular Insights into Epigenetics and Cannabinoid Receptors" Biomolecules 12, no. 11: 1560. https://doi.org/10.3390/biom12111560
APA StyleBasavarajappa, B. S., & Subbanna, S. (2022). Molecular Insights into Epigenetics and Cannabinoid Receptors. Biomolecules, 12(11), 1560. https://doi.org/10.3390/biom12111560







