Using GPT-3 to Build a Lexicon of Drugs of Abuse Synonyms for Social Media Pharmacovigilance
Abstract
:1. Introduction
- We introduce a novel method to repeatedly query GPT-3 for drug synonyms and filter the generated terms to create a lexicon enriched for likely synonyms, all in an automated fashion. We make the code for the method publicly available to build similar lexicons, facilitating interpretable pharmacovigilance on messy, casually-written social media data that does not require training a new large machine learning model;
- We present a lexicon of GPT-3 synonyms for 98 drugs of abuse, including 22 widely-discussed drugs of abuse, which can be used to easily flag text likely to be related to drug abuse from a large corpus of informal language in an interpretable manner;
- Finally, we also demonstrate of the capabilities of GPT-3, and similar models, for practical contributions to pharmacovigilance.
2. Materials and Methods
2.1. Datasets
2.1.1. RedMed
2.1.2. Drugs of Abuse
2.1.3. Widely-Discussed Drugs of Abuse
2.2. External Models
2.2.1. GPT-3
2.2.2. Google Search API
2.3. Terminology
2.4. Methods
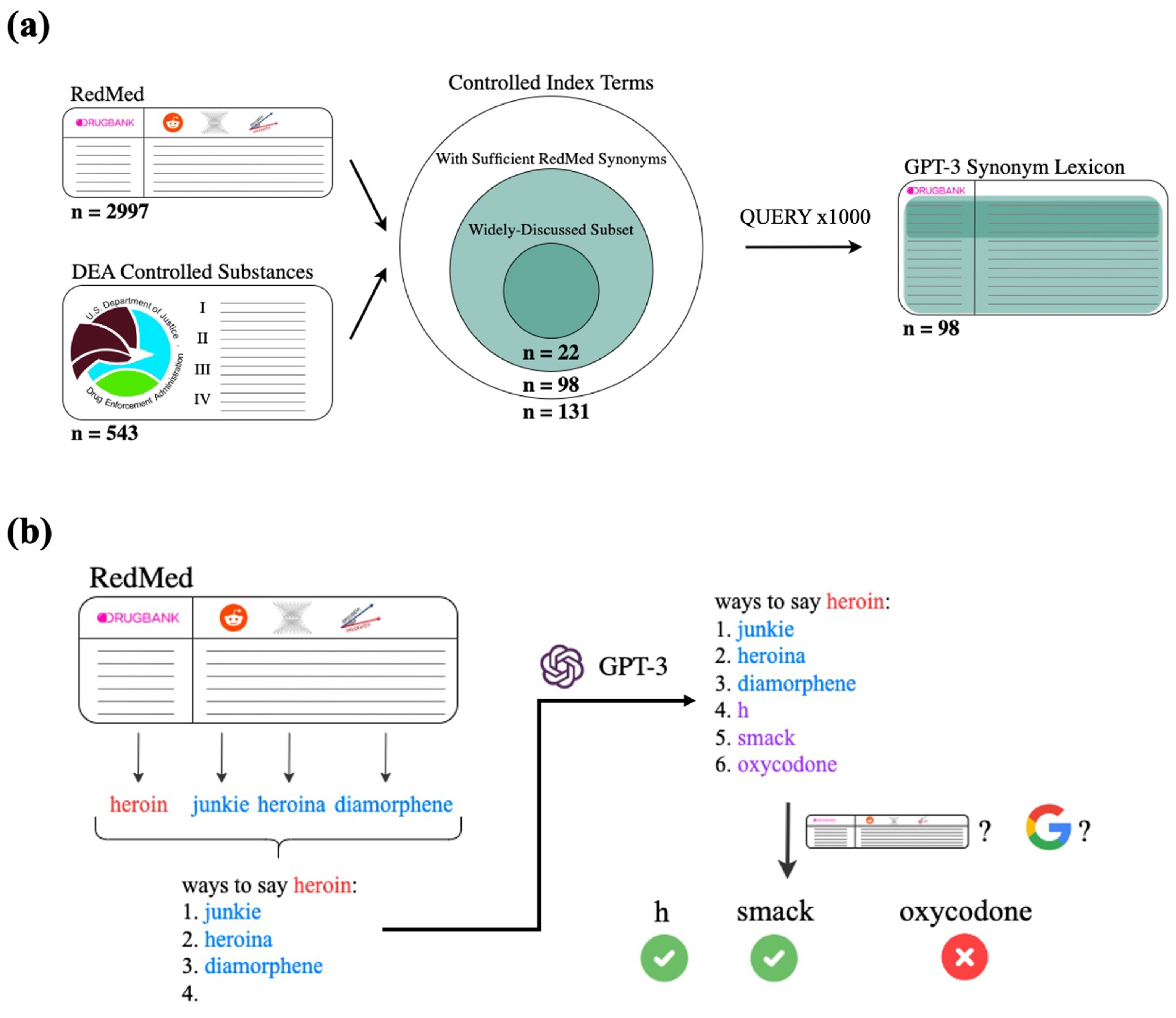
2.4.1. Overview of Query Pipeline
2.4.2. GPT-3 Prompt Templates
“ways to say [index term]:
1. [RedMed synonym 1]
2. [RedMed synonym 2]
3. [RedMed synonym 3]
4.”
“these are not synonyms for [index term]:
1. [counterexample 1]
2. [counterexample 2]
3. [counterexample 3]
4. [counterexample 4]
but these are synonyms for [index term]:
1. [RedMed synonym 1]
2. [RedMed synonym 2]
3.”
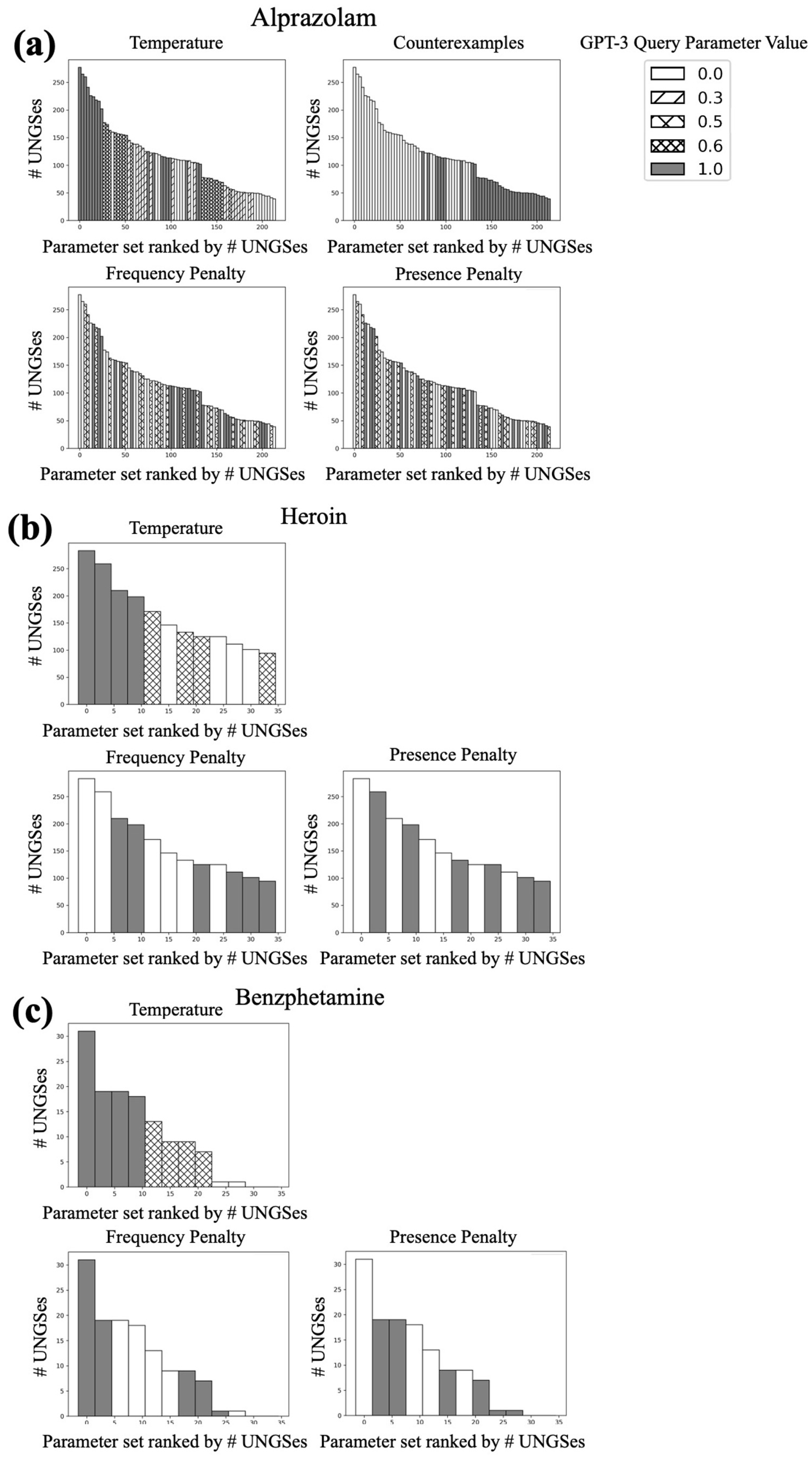
2.4.3. GPT-3 Parameter Search
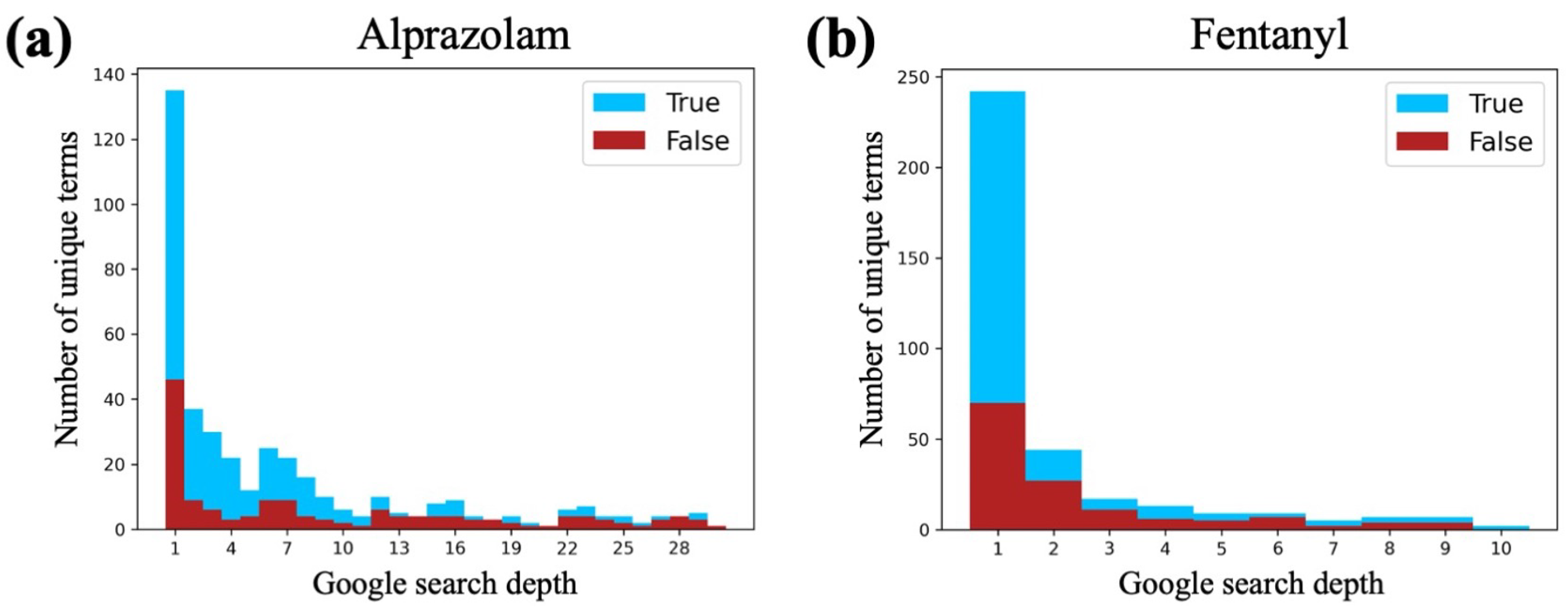
2.4.4. Google Filter
2.4.5. Drug Name Filter
2.4.6. Final Pipeline Parameters
2.4.7. Manual Labeling
2.4.8. Evaluation Criteria
3. Results
3.1. Parameter Search
3.2. Google Search Depth Analysis
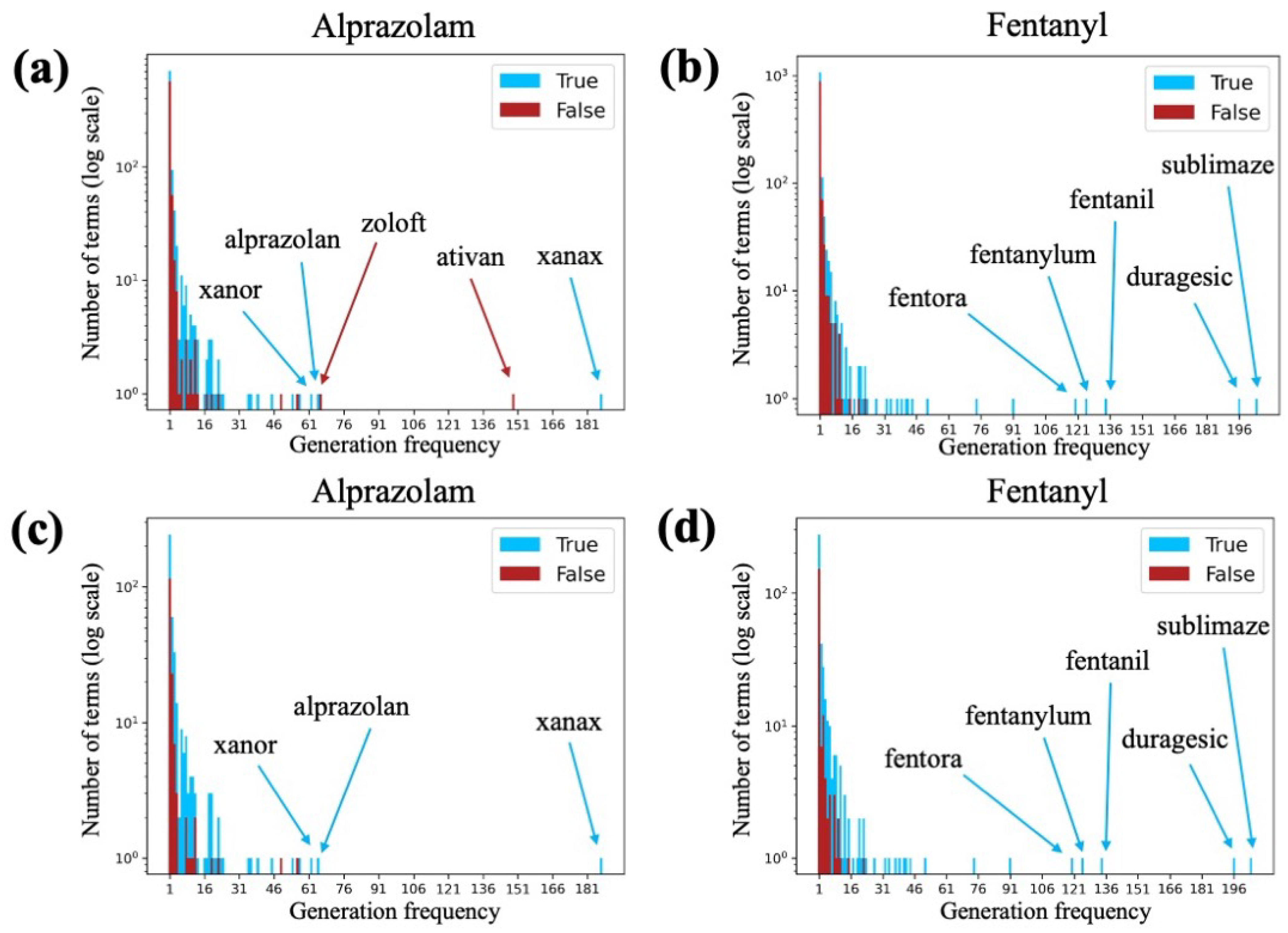
3.3. Generation Frequency Analysis
3.4. Pipeline Performance
3.5. Drugs of Abuse Lexicon
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GPT-3 | Generative Pretrained Transformer 3 |
| WHO | World Health Organization |
| EMA | European Medicines Agency |
| FDA | U.S. Food and Drug Administration |
| CDC | U.S. Centers for Disease Control and Prevention |
| NIH | U.S. National Institutes of Health |
| DEA | U.S. Drug Enforcement Administration |
| HHS | U.S. Department of Health and Human Services |
| FAERS | FDA Adverse Event Reporting System |
| NHANES | National Health and Nutrition Examination Survey |
| NDEWS | National Drug Early Warning System |
| NFLIS | National Forensic Laboratory Information System |
| NSDUH | National Survey on Drug Use and Health |
| NLP | Natural Language Processing |
| EHR | Electronic Health Record |
| AKDT | Associated Known Drug Term |
| API | Application Programming Interface |
| UNGS | Unique Novel GPT-3 Synonym |
| TP | True Positive |
| FP | False Positive |
| FN | False Negative |
Appendix A

References
- Lyden, J.; Binswanger, I.A. The United States opioid epidemic. Semin. Perinatol. 2019, 43, 123. [Google Scholar] [CrossRef] [PubMed]
- Ciccarone, D. The Rise of Illicit Fentanyls, Stimulants and the Fourth Wave of the Opioid Overdose Crisis. Curr. Opin. Psychiatry 2021, 34, 344. [Google Scholar] [CrossRef]
- CDC. National Center for Health Statistics. 2021. Wide-Ranging Nnline Data for Epidemiologic Research (WONDER). Available online: http://wonder.cdc.gov (accessed on 19 December 2022).
- Beninger, P. Pharmacovigilance: An Overview. Clin. Ther. 2018, 40, 1991–2004. [Google Scholar] [CrossRef] [Green Version]
- Throckmorton, D.C.; Gottlieb, S.; Woodcock, J. The FDA and the Next, Wave of Drug Abuse — Proactive Pharmacovigilance. N. Engl. J. Med. 2018, 379, 205–207. [Google Scholar] [CrossRef] [PubMed]
- Stokes, A.; Berry, K.M.; Hempstead, K.; Lundberg, D.J.; Neogi, T. Trends in Prescription Analgesic Use Among Adults With Musculoskeletal Conditions in the United States, 1999–2016. JAMA Netw. Open 2019, 2, e1917228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tringale, K.R.; Huynh-Le, M.P.; Salans, M.; Marshall, D.C.; Shi, Y.; Hattangadi-Gluth, J.A. The role of cancer in marijuana and prescription opioid use in the United States: A population-based analysis from 2005 to 2014. Cancer 2019, 125, 2242–2251. [Google Scholar] [CrossRef] [PubMed]
- Veronin, M.A.; Schumaker, R.P.; Dixit, R.R.; Elath, H. Opioids and frequency counts in the US Food and Drug Administration Adverse Event Reporting System (FAERS) database: A quantitative view of the epidemic. Drug Healthc. Patient Saf. 2019, 11, 65. [Google Scholar] [CrossRef] [Green Version]
- Elmore, A.L.; Omofuma, O.O.; Sevoyan, M.; Richard, C.; Liu, J. Prescription opioid use among women of reproductive age in the United States: NHANES, 2003–2018. Prev. Med. 2021, 153, 106846. [Google Scholar] [CrossRef]
- Robert, M.; Jouanjus, E.; Khouri, C.; Sam-Laï, N.F.; Revol, B. The opioid epidemic: A worldwide exploratory study using the WHO pharmacovigilance database. Addiction 2022. [Google Scholar] [CrossRef]
- Chiappini, S.; Vickers-Smith, R.; Guirguis, A.; Corkery, J.M.; Martinotti, G.; Harris, D.R.; Schifano, F. Pharmacovigilance Signals of the Opioid Epidemic over 10 Years: Data Mining Methods in the Analysis of Pharmacovigilance Datasets Collecting Adverse Drug Reactions (ADRs) Reported to EudraVigilance (EV) and the FDA Adverse Event Reporting System (FAERS). Pharmaceuticals 2022, 15, 675. [Google Scholar] [CrossRef]
- Inoue, K.; Ritz, B.; Arah, O.A. Causal Effect of Chronic Pain on Mortality Through Opioid Prescriptions: Application of the Front-Door Formula. Epidemiology 2022, 33, 572. [Google Scholar] [CrossRef] [PubMed]
- Marwitz, K.K.; Noureldin, M. A descriptive analysis of concomitant opioid and benzodiazepine medication use and associated adverse drug events in United States adults between 2009 and 2018. Explor. Res. Clin. Soc. Pharm. 2022, 5, 100130. [Google Scholar] [CrossRef] [PubMed]
- LePendu, P.; Iyer, S.V.; Bauer-Mehren, A.; Harpaz, R.; Ghebremariam, Y.T.; Cooke, J.P.; Shah, N.H. Pharmacovigilance using Clinical Text. AMIA Summits Transl. Sci. Proc. 2013, 2013, 109. [Google Scholar] [PubMed]
- Harpaz, R.; Callahan, A.; Tamang, S.; Low, Y.; Odgers, D.; Finlayson, S.; Jung, K.; LePendu, P.; Shah, N.H. Text mining for adverse drug events: The promise, challenges, and state of the art. Drug Saf. 2014, 37, 777–790. [Google Scholar] [CrossRef]
- Boland, M.R.; Tatonetti, N.P. Are All Vaccines Created Equal? Using Electronic Health Records to Discover Vaccines Associated With Clinician-Coded Adverse Events. AMIA Summits Transl. Sci. Proc. 2015, 2015, 196. [Google Scholar]
- Luo, Y.; Thompson, W.K.; Herr, T.M.; Zeng, Z.; Berendsen, M.A.; Jonnalagadda, S.R.; Carson, M.B.; Starren, J. Natural Language Processing for EHR-Based Pharmacovigilance: A Structured Review. Drug Saf. 2017, 40, 1075–1089. [Google Scholar] [CrossRef]
- Ward, P.J.; Rock, P.J.; Slavova, S.; Young, A.M.; Bunn, T.L.; Kavuluru, R. Enhancing timeliness of drug overdose mortality surveillance: A machine learning approach. PLoS ONE 2019, 14, e0223318. [Google Scholar] [CrossRef] [Green Version]
- Ward, P.J.; Young, A.M.; Slavova, S.; Liford, M.; Daniels, L.; Lucas, R.; Kavuluru, R. Deep Neural Networks for Fine-Grained Surveillance of Overdose Mortality. Am. J. Epidemiol. 2022, 192, 257–266. [Google Scholar] [CrossRef]
- Kazemi, D.M.; Borsari, B.; Levine, M.J.; Dooley, B. Systematic review of surveillance by social media platforms for illicit drug use. J. Public Health 2017, 39, 763–776. [Google Scholar] [CrossRef] [Green Version]
- Leaman, R.; Wojtulewicz, L.; Sullivan, R.; Skariah, A.; Yang, J.; Gonzalez, G. Toward Internet-Age Pharmacovigilance: Extracting Adverse Drug Reactions from User Posts to Health-Related Social Networks. In Proceedings of the of the 2010 Workshop on Biomedical Natural Language Processing, Uppsala, Sweden, 15 July 2010; pp. 117–125. [Google Scholar]
- O’Connor, K.; Pimpalkhute, P.; Nikfarjam, A.; Ginn, R.; Smith, K.L.; Gonzalez, G. Pharmacovigilance on Twitter? Mining Tweets for Adverse Drug Reactions. AMIA Annu. Symp. Proc. 2014, 2014, 924. [Google Scholar]
- Eshleman, R.; Singh, R. Leveraging graph topology and semantic context for pharmacovigilance through Twitter-streams. BMC Bioinform. 2016, 17. [Google Scholar] [CrossRef] [Green Version]
- Cocos, A.; Fiks, A.G.; Masino, A.J. Deep learning for pharmacovigilance: Recurrent neural network architectures for labeling adverse drug reactions in Twitter posts. J. Am. Med Inform. Assoc. JAMIA 2017, 24, 813. [Google Scholar] [CrossRef]
- Lardon, J.; Bellet, F.; Aboukhamis, R.; Asfari, H.; Souvignet, J.; Jaulent, M.C.; Beyens, M.N.; Lillo-LeLouët, A.; Bousquet, C. Evaluating Twitter as a complementary data source for pharmacovigilance. Expert Opin. Drug Saf. 2018, 17, 763–774. [Google Scholar] [CrossRef]
- Farooq, H.; Niaz, J.S.; Fakhar, S.; Naveed, H. Leveraging digital media data for pharmacovigilance. AMIA Annu. Symp. Proc. 2020, 2020, 442. [Google Scholar]
- Magge, A.; Tutubalina, E.; Miftahutdinov, Z.; Alimova, I.; Dirkson, A.; Verberne, S.; Weissenbacher, D.; Gonzalez-Hernandez, G. DeepADEMiner: A deep learning pharmacovigilance pipeline for extraction and normalization of adverse drug event mentions on Twitter. J. Am. Med Inform. Assoc. 2021, 28, 2184–2192. [Google Scholar] [CrossRef]
- Pierce, C.E.; Bouri, K.; Pamer, C.; Proestel, S.; Rodriguez, H.W.; Le, H.V.; Freifeld, C.C.; Brownstein, J.S.; Walderhaug, M.; Edwards, I.R.; et al. Evaluation of Facebook and Twitter Monitoring to Detect Safety Signals for Medical Products: An Analysis of Recent FDA Safety Alerts. Drug Saf. 2017, 40, 317–331. [Google Scholar] [CrossRef] [Green Version]
- Caster, O.; Dietrich, J.; Kürzinger, M.L.; Lerch, M.; Maskell, S.; Norén, G.N.; Tcherny-Lessenot, S.; Vroman, B.; Wisniewski, A.; van Stekelenborg, J. Assessment of the Utility of Social Media for Broad-Ranging Statistical Signal Detection in Pharmacovigilance: Results from the WEB-RADR Project. Drug Saf. 2018, 41, 1355. [Google Scholar] [CrossRef] [Green Version]
- Hussain, Z.; Sheikh, Z.; Tahir, A.; Dashtipour, K.; Gogate, M.; Sheikh, A.; Hussain, A. Artificial Intelligence–Enabled Social Media Analysis for Pharmacovigilance of COVID-19 Vaccinations in the United Kingdom: Observational Study. JMIR Public Health Surveill. 2022, 8, e32543. [Google Scholar] [CrossRef]
- Natter, J.; Michel, B. Memantine misuse and social networks: A content analysis of Internet self-reports. Pharmacoepidemiol. Drug Saf. 2020, 29, 1189–1193. [Google Scholar] [CrossRef]
- Lavertu, A.; Hamamsy, T.; Altman, R.B. Quantifying the Severity of Adverse Drug Reactions Using Social Media: Network Analysis. J. Med. Internet Res. 2021, 23, e27714. [Google Scholar] [CrossRef]
- Preiss, A.; Baumgartner, P.; Edlund, M.J.; Bobashev, G.V. Using Named Entity Recognition to Identify Substances Used in the Self-medication of Opioid Withdrawal: Natural Language Processing Study of Reddit Data. JMIR Form. Res. 2022, 6, e33919. [Google Scholar] [CrossRef] [PubMed]
- Tasnim, S.; Hossain, M.; Mazumder, H. Impact of Rumors and Misinformation on COVID-19 in Social Media. J. Prev. Med. Public Health 2020, 53, 171. [Google Scholar] [CrossRef] [Green Version]
- Suarez-Lledo, V.; Alvarez-Galvez, J. Prevalence of Health Misinformation on Social Media: Systematic Review. J. Med. Internet Res. 2021, 23, e17187. [Google Scholar] [CrossRef] [PubMed]
- Convertino, I.; Ferraro, S.; Blandizzi, C.; Tuccori, M. The usefulness of listening social media for pharmacovigilance purposes: A systematic review. Expert Opin. Drug Saf. 2018, 17, 1081–1093. [Google Scholar] [CrossRef] [PubMed]
- Tricco, A.C.; Zarin, W.; Lillie, E.; Jeblee, S.; Warren, R.; Khan, P.A.; Robson, R.; Pham, B.; Hirst, G.; Straus, S.E. Utility of social media and crowd-intelligence data for pharmacovigilance: A scoping review. BMC Med. Inform. Decis. Mak. 2018, 18. [Google Scholar] [CrossRef] [Green Version]
- Pappa, D.; Stergioulas, L.K. Harnessing social media data for pharmacovigilance: A review of current state of the art, challenges and future directions. Int. J. Data Sci. Anal. 2019, 8, 113–135. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.Y.; Lee, Y.S.; Kim, D.H.; Lee, H.S.; Yang, B.R.; Kim, M.G. The Use of Social Media in Detecting Drug Safety–Related New Black Box Warnings, Labeling Changes, or Withdrawals: Scoping Review. JMIR Public Health Surveill. 2021, 7, e30137. [Google Scholar] [CrossRef]
- Lavertu, A.; Altman, R.B. RedMed: Extending drug lexicons for social media applications. J. Biomed. Inform. 2019, 99, 103307. [Google Scholar] [CrossRef]
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient Estimation of Word Representations in Vector Space. arXiv 2013, arXiv:1301.378. [Google Scholar]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. NUcleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Brown, T.B.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; et al. Language Models are Few-Shot Learners. In Proceedings of the NeurIPS 2020, Virtual. 6–12 December 2020. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Łukasz, K.; Polosukhin, I. Attention Is All You Need. In Proceedings of the NeurIPS 2017, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Korngiebel, D.M.; Mooney, S.D. Considering the possibilities and pitfalls of Generative Pre-trained Transformer 3 (GPT-3) in healthcare delivery. NPJ Digit. Med. 2021, 4, 93. [Google Scholar] [CrossRef]
- Sezgin, E.; Sirrianni, J.; Linwood, S.L. Operationalizing and Implementing Pretrained, Large Artificial Intelligence Linguistic Models in the US Health Care System: Outlook of Generative Pretrained Transformer 3 (GPT-3) as a Service Model. JMIR Med. Inform. 2022, 10, e32875. [Google Scholar] [CrossRef]
- Nath, S.; Marie, A.; Ellershaw, S.; Korot, E.; Keane, P.A.; Pearse, D.; Keane, A. New meaning for NLP: The trials and tribulations of natural language processing with GPT-3 in ophthalmology. Br. J. Ophthalmol. 2022, 106, 889–892. [Google Scholar] [CrossRef]
- Mcguffie, K.; Newhouse, A. The radicalization risks of GPT-3 and advanced neural language models. arXiv 2020, arXiv:2009.06807. [Google Scholar]
- Abid, A.; Farooqi, M.; Zou, J. Persistent Anti-Muslim Bias in Large Language Models. arXiv 2021, arXiv:2101.05783. [Google Scholar]
- U.S. Drug Enforcement Administration, Controlled Substances—Alphabetical Order. Available online: https://www.deadiversion.usdoj.gov/schedules/orangebook/c_cs_alpha.pdf (accessed on 25 July 2022).
- Park, A.; Conway, M. Tracking Health Related Discussions on Reddit for Public Health Applications. AMIA Annu. Symp. Proc. 2017, 2017, 1362. [Google Scholar]
- Adams, N.; Artigiani, E.E.; Wish, E.D. Choosing Your Platform for Social Media Drug Research and Improving Your Keyword Filter List. J. Drug Issues 2019, 49, 477–492. [Google Scholar] [CrossRef]
- OpenAI. ChatGPT: Optimizing Language Models for Dialogue. 2022. Available online: https://openai.com/blog/chatgpt/ (accessed on 16 December 2022).




| Term | Abbreviation | Definition |
|---|---|---|
| Associated known drug term | AKDT | As defined in [40], known terms used synonymously for a given drug, most often brand names. |
| Controlled index term | - | A controlled substance that is an index term. |
| Controlled substance | - | A substance (i.e., drug) that is deemed to have a high potential for abuse by the DEA and is therefore controlled. |
| Generated term | - | See GPT-3 generated term. |
| GPT-3 generated term | - | A term generated from a GPT-3 query as a candidate synonym for the corresponding index term used in the prompt. |
| GPT-3 synonym | - | A GPT-3 generated term that has been automatically labeled as a synonym following a filtering scheme. |
| Index term | - | The identifying term of a drug as indexed in RedMed; also the generic name of a drug as indicated in DrugBank. |
| Novel GPT-3 synonym | - | A GPT-3 synonym that is not already present in RedMed as a RedMed synonym. |
| Non-synonym | - | A generated term that has been manually labeled as not synonymous for the corresponding queried index term. |
| RedMed synonym | - | A term listed in RedMed as synonymous for a given index term. |
| Synonym | - | A generated term that has been manually labeled as synonymous for the corresponding queried index term. |
| Unique novel GPT-3 synonym | UNGS | Equivalent to a novel GPT-3 synonym but specifying that each unique novel GPT-3 synonym is only counted once no matter how many times it has been generated. |
| Widely-discussed | - | Specifying that a drug appears relatively more frequently on Reddit, suggesting higher rates of online discussion, more synonymous terms, and potentially greater interest for pharmacovigilance. |
| Index Term | GPT-3 Synonym Criteria | Precision | Recall | F1 Score | F2 Score |
|---|---|---|---|---|---|
| Alprazolam | All generated terms | 0.264 | 1.000 | 0.418 | 0.642 |
| Fentanyl | All generated terms | 0.220 | 1.000 | 0.361 | 0.585 |
| Alprazolam | All RedMed terms | 1.000 | 0.178 | 0.302 | 0.213 |
| Fentanyl | All RedMed terms | 1.000 | 0.115 | 0.206 | 0.140 |
| Alprazolam | Drug name filter | 0.285 | 0.996 | 0.443 | 0.664 |
| Fentanyl | Drug name filter | 0.232 | 1.000 | 0.377 | 0.602 |
| Alprazolam | Drug name & frequency filters | 0.567 | 0.487 | 0.524 | 0.501 |
| Fentanyl | Drug name & frequency filters | 0.521 | 0.465 | 0.491 | 0.475 |
| Alprazolam | Drug name & Google filters | 0.698 | 0.859 | 0.770 | 0.821 |
| Fentanyl | Drug name & Google filters | 0.568 | 0.793 | 0.662 | 0.735 |
| Alprazolam | Drug name, frequency, & Google filters | 0.859 | 0.431 | 0.574 | 0.479 |
| Fentanyl | Drug name, frequency, & Google filters | 0.770 | 0.395 | 0.522 | 0.438 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carpenter, K.A.; Altman, R.B. Using GPT-3 to Build a Lexicon of Drugs of Abuse Synonyms for Social Media Pharmacovigilance. Biomolecules 2023, 13, 387. https://doi.org/10.3390/biom13020387
Carpenter KA, Altman RB. Using GPT-3 to Build a Lexicon of Drugs of Abuse Synonyms for Social Media Pharmacovigilance. Biomolecules. 2023; 13(2):387. https://doi.org/10.3390/biom13020387
Chicago/Turabian StyleCarpenter, Kristy A., and Russ B. Altman. 2023. "Using GPT-3 to Build a Lexicon of Drugs of Abuse Synonyms for Social Media Pharmacovigilance" Biomolecules 13, no. 2: 387. https://doi.org/10.3390/biom13020387
APA StyleCarpenter, K. A., & Altman, R. B. (2023). Using GPT-3 to Build a Lexicon of Drugs of Abuse Synonyms for Social Media Pharmacovigilance. Biomolecules, 13(2), 387. https://doi.org/10.3390/biom13020387







