1. Introduction
Parkinson’s disease (PD), a long-term neurodegenerative disorder affecting the human motor system, results in many motor and non-motor characteristics [
1]. It is considered one of the most common movement disorders among individuals over 60 years of age [
2]. PD has a Relative Risk (RR) of death of 2.3 (1.8 to 3.0) [
3]. PD can be detected by observing changes in behavioral patterns, as well as by observing rigidity, cognitive impairment, bradykinesia, tremors, and postural instability [
4]. PD is currently incurable, but treatment following early diagnosis can improve and relieve the symptoms. Most people experience a normal life expectancy of another 7 to 14 years after diagnosing PD [
5]. However, PD has a more than 25% rate of misdiagnosis [
6]. More accurate detection of PD can increase the life expectancy for a diagnosed PD patient by preventing complications and maintaining a high quality of life with the necessary pharmacological and surgical intervention [
7].
In order to detect PD at an early stage, many health informatics systems, including telediagnosis and telemonitoring systems, have been developed for current pharmacological therapeutics [
8]. Researchers are concentrating on identifying biological markers for the detection phase. At present, the application of Machine Learning (ML) is thriving in the field of prediction; hence, it is widely used in PD diagnosis. Neuroimaging modalities, including feature extraction from the processing of scanned data such as MRI images using different ML techniques, are paving a promising way towards detecting PD [
9,
10,
11]. These health informatics systems aim to reduce the discommoding physical visits to clinics for clinical examination, which, as a result, will reduce the task of health workers and clinicians [
12,
13,
14,
15].
There are many symptoms that appear in PD patients, including posture and balance deficiencies, dysphonia (change in speech and articulation), and slowed movement, etc. Among these symptoms is vocal dysfunction, which results in vocal instability, loudness, and damaged vocal quality. Therefore, the early detection of PD can be done by analyzing speech signals, as 90% of PD patients face vocal problems in the incipient stage of the disease [
16]. Consequently, the recent focus of PD detection research emphasizes the vocal disorders of patients. In recent studies, clinical features from the speech of PD patients were extracted and fed into a classification model with the help of various speech signal processing algorithms. Based on speech recordings, this telemonitoring study mapped the vocal features of PD to a clinical evaluation system that predicted the possibility of PD in patients. Moreover, the collection of speech data was a non-invasive process, which made the data easy to collect and subsequently provide as the input of the telediagnosis system.
ML techniques including Artificial Neural Networks (ANN) [
17], K-Nearest Neighbors (KNN) [
18], Random Forest (RF) [
19], and Extreme Gradient Boosting (XGBoost) [
20] have been used in PD classification based on patient vocal disorders. However, the success rate of accurate detection depends on the quality of data, on the relevance of the features extracted from them, and on the associated ML models. Many recent studies have been conducted on a publicly available dataset that consists of the sound measurements of 8 healthy and 23 PD-affected instances, aggregating 195 data samples [
21]. Another publicly available dataset includes the data of 20 PD patients and 20 healthy individuals [
12]. Both datasets consist of some common features extracted from the speech signals, including vocal fundamental frequency, measures of the ratio of the noise-to-tonal components, measures of variation in amplitude, measures of variation in fundamental frequency, etc. Since a good number of the studies regarding PD classification from vocal features are conducted with these datasets, the features extracted from these datasets are referred to as the baseline features. Other features are also used to detect PD, such as Mel-frequency Cepstral Coefficients (MFCC) and Signal-to-Noise Ratio (SNR) [
22]. In a recent study, the effectiveness of vocal features was analyzed [
15]. Although slow movement, tremors, inertia, and balance deficiency are among the symptoms of PD, vocal and speech signal processing is widely used as they can be easily tracked by the changes of speech along with other symptoms’ data, as formerly stated, from wearable sensors.
Sharma and Giri extracted different features from voice signals, including MFCC, jitter, shimmer features, pitch, and glottal pulse [
23]. These feature values of a PD patient show higher fluctuations and variance than those of a normal person. Tunable Q-factor Wavelet Transform (TQWT) was introduced in a recent study by Sakar et al. [
16], along with other features, to detect PD patients from their vocal signals. The same dataset was being experimented in a study using Deep Learning techniques, where two frameworks consisting of Convolutional Neural Networks (CNN) were applied [
24]. However, properly training Deep Neural Networks (DNN) to converge requires very large datasets and also takes substantial training time in order to search the parameter space. In addition, dividing all the features into different sets, a feature that is more relevant and important in the classification process, is treated in the same way as other less important features of the same set, which decreases the accuracy of detecting a PD patient.
Bouchikhi et al. [
25] proposed a model with an SVM as a classifier and Relief-F as a feature selection method. In their study, the feature set was reduced from 22 to 10. Subsequently, the SVM classifier with a 10-fold cross-validation method showed that Relief-F had an accuracy rate of 96.88%. The dataset contained 195 voice samples. However, it was empirically and theoretically proved that the performance of core relief-based algorithms (RBA) decreases drastically as the number of irrelevant features becomes enormous [
26]. This is because as the number of features increases, Relief’s computation of neighbors and weights becomes excessively random, which is an example of curse dimensionality. Additionally, [
27] found that RBAs are vulnerable to noise interfering with the section of nearest neighbors.
Hemmerling and Sztahó [
28], on the other hand, proposed the use of PCA with a non-linear SVM to identify PD patients, and their methods show a classification accuracy rate of 93.43%. However, the dataset used in their study was comparatively small. Small datasets have several disadvantages, leading to lower precision in the prediction and lower power. They also pose a greater risk as they may compare the classes unfairly, even in circumstances where the data is from a randomized trial.
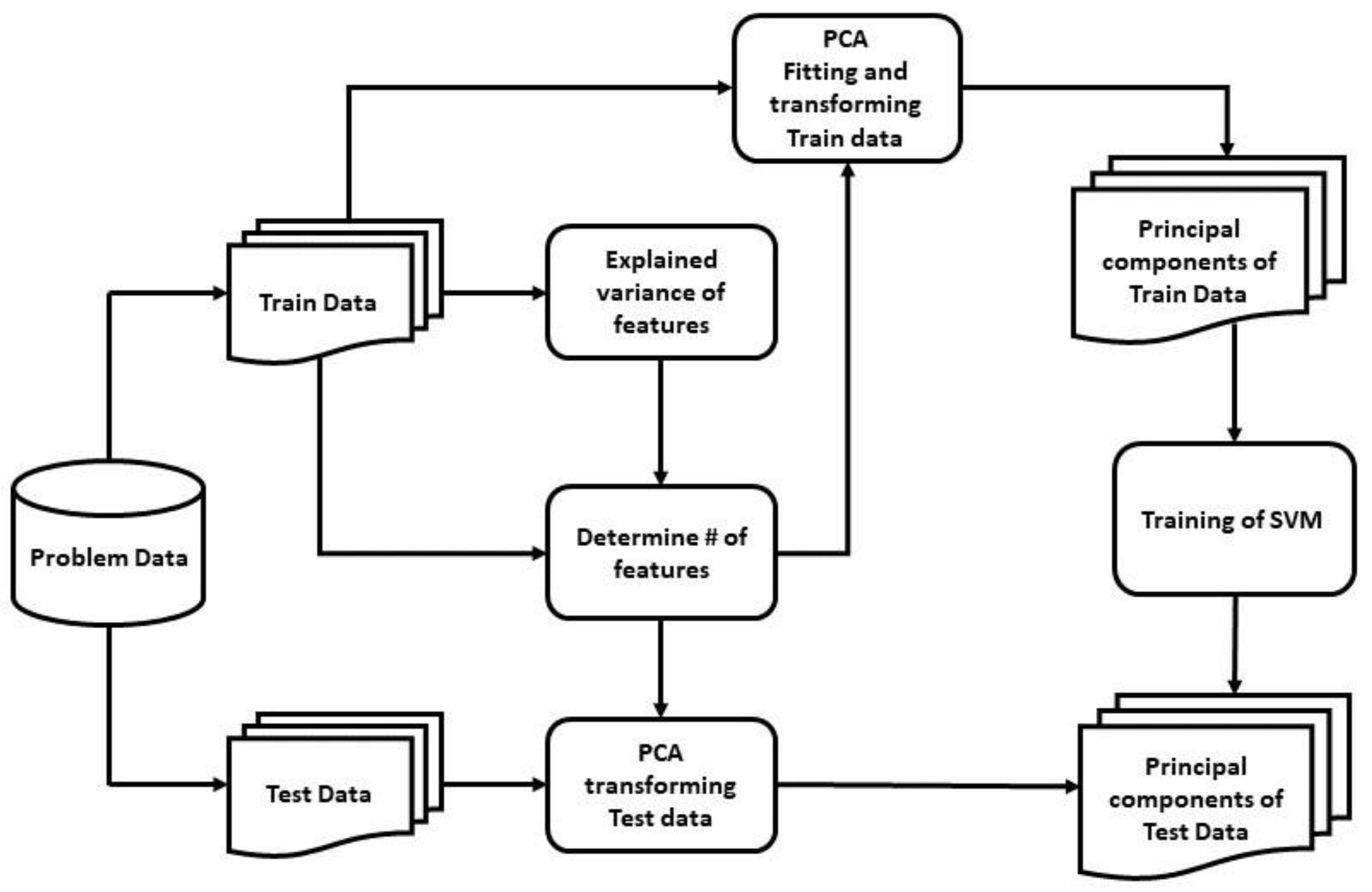
Our study adopted two different feature extraction techniques, including Principal Component Analysis (PCA) [
29] and a novel deeper Sparse Autoencoder (SAE) [
30], where the PCA is widely used when the features are linearly related and our novel SAE is capable of working well with non-linearly related features in order to reduce the feature set dimensionality and take into account the most important and relevant information for classification. Moreover, feeding the model irrelevant and high dimensional data risks over-fitting. In order to classify between a PD patient and a healthy individual, the Support Vector Machine [
31,
32] is employed as a powerful classification method that exceeds other techniques with fewer subjects. When the number of features is high, SVM is more efficient at classification task in high dimensional spaces with less training data, delivering a unique solution surpassing the Neural Networks in the case of convex optimality problems. This study aimed to eliminate the drawbacks of both Gunduz [
24] and Sakar et al. [
16] by applying two hybrid models integrating PCA-SVM and SAE-SVM.
The rest of the paper is organized as follows:
Section 2 summarizes the related work conducted in this domain,
Section 3 contains the description of the dataset,
Section 4 describes the methodologies that are used in this study,
Section 5 states the experimental results and comparison with different traditional and state-of-the-art disciplines, and
Section 6 briefly draws the findings and future directions of research.
2. Related Works
In this section, various recent studies on PD classification using machine learning algorithms and deep learning methods have been summarized.
In [
33], Karimi Rouzbahani and Daliri used voice signals for PD detection. Parameters such as pitch, jitter, fundamental frequency, shimmer, and various statistical measures based on these parameters were used as the input of the proposed predictive model. The harmonics-to-noise ratio and the noise-to-harmonics ratio were also extracted using estimates of signal-to-noise by calculation of the autocorrelation of each cycle. In their study, one of every two features which resulted in a correlation rate of over 95% was removed. Several feature selection methods, such as Receiver Operating Characteristics (ROC) curves,
t-test, and Fisher’s Discriminant Ratio (FDR), were utilized to identify the informative features among the whole feature set. The number of features was incremented one by one. Afterwards, the prioritized features were fed into an SVM classifier. The highest performance was achieved using three classifiers at hand and a combination of the seven most prior features. The selected features were used to train the SVM, KNN, and discrimination-function-based (DBF) classifiers. Among these classifiers, KNN showed the best performance with an accuracy rate of 93.82%. KNN has also shown good performance in other performance matrices such as error rate, sensitivity, and specificity.
Ma et al. [
34] proposed a novel hybrid method integrating subtractive clustering features weighting (SCFW) and kernel-based extreme learning machine (KELM) for the diagnosis of PD patients. SCFW, a data-preprocessing tool, is used to decrease the variance in the dataset. The output of the SCFW further improved the accuracy of the KELM classifier. The efficiency of the proposed method was justified in terms of accuracy, specificity, sensitivity, Area under the ROC Curve (AUC), kappa statistic value, and f-measure. The proposed method outperformed (via a 10-fold cross-validation scheme) the SVM-based, ELM-based, KNN-based, and other methods with a classification accuracy of 99.49%.
Zuo et al. [
35] presented an effective method for the diagnosis of PD patients using particle swarm optimization (PSO) enhanced fuzzy k-nearest neighbor (FKNN). PSO-FKNN uses both the continuous and binary versions of PSO to achieve parameter optimization and feature selection concurrently. The continuous PSO adaptively specifies the fuzzy strength parameter m in and the neighborhood size k in FKNN, whereas the most discerning subset of features in the dataset is chosen by the binary PSO. This PSO-FKNN model was justified in terms of the accuracy, specificity, sensitivity, and the AUC of the ROC curve. The proposed model achieved a mean accuracy rate of 97.47% via a 10-fold cross-validation analysis.
Sharma and Giri [
23] applied three types of classifiers based on MLP, KNN, and SVM to diagnose PD patients. Among these, SVM with an RBF kernel showed the best result with a total classification accuracy rate of 85.294%.
Parisi et al. [
36] proposed an artificial intelligent classifier using Multilayer Perceptron (MLP) and Lagrangian Support Vector Machine (LSVM). The importance scores of the features were assigned by the MLP with custom cost functions, which included both the AUC score and the accuracy. The MLP provided the 20 most important features with high importance scores, which were then fed into the LSVM classifier. The proposed method achieved an accuracy rate of 100% and was compared with other similar studies.
For the first time, Sakar et al. [
16] introduced tunable Q-factor wavelet transform (TQWT) to vocal signals for the diagnosis of PD patients. The feature subsets obtained from the dataset were given as input in various classifiers, and the study showed that TQWT-based features tend to achieve better results than the other popular voice features used in PD classification. In their study, a combination of TQWT plus Mel-frequency cepstral coefficients (MFCC) plus Concat showed the best performance in terms of all metrics and among all classifiers with an accuracy rate of 85.7%, an F1-score of 0.84, and an MCC value of 0.59.
Gunduz [
24] used the same dataset used in Sakar et al. [
16] to diagnose PD patients using Deep Learning Techniques, where two frameworks consisting of CNN were applied. Both of the studies used the features as subsets and estimated the accuracy by a combination of input into the ML models. Gunduz [
24] showed a higher accuracy of 86.9%, an F1-score of 0.910, and an MCC value of 0.632.
Caliskan et al. [
37] proposed a PD detection model, which is a DNN classifier consisting of a stacked autoencoder used for obtaining inherent information within the vocal features. Their proposed model was compared with several other state-of-the-art ML models, and it was concluded that the DNN classifier was convenient in the diagnosis of PD patients. However, DNN requires plenty of data to be suitably trained to converge and also takes a lot of training time in order to search the parameter space. Moreover, their study focused only on feature extraction by redundancy removal while classifying PD patients.
In another study, Wroge et al. [
38] performed the diagnosis of PD patients using DNN. A mobile application was used to collect the data of PD patients and non-PD patients. Two types of feature sets were obtained from the collected data. The first one was the Audio-Visual Emotion recognition Challenge (AVEC), which had dimensions up to 2200, and the second features set contained 60 features that were set up using MFCC. The features were given as input in a three-layered DNN and other AI classifiers. The results showed that DNN had the highest accuracy rate of 85% compared to the average clinical diagnosis accuracy rate of 73.8%.
The above-mentioned studies are based on vocal features as an important factor for detecting PD patients. Apart from these, several other studies have also been performed which extracts features from different datasets, e.g., wearable sensors [
39], electroencephalogram (EEG) [
40], and smart pens [
41].
3. Dataset
The dataset was obtained from the University of California-Irvine (UCI) Machine Learning repository and was used in [
16,
24] before the present study.
Table 1 contains the details of the dataset. The dataset has an imbalance regarding the number of men and women (a ratio of 23:41) and healthy individuals and PD patients (a ratio of 107:81). PD is 1.5 times more common in men, and along with that, motor progression is more aggressive in men than in women. Nevertheless, there are no significant differences in terms of demographic variables [
42].
As speech anomalies are one of the key effects that have been seen in PD patients, vocal features and speech signal attributes have been used successfully to assess PD. The traditional features mostly used in PD detection are the fundamental frequency parameters, Recurrence Period Density Entropy (RPDE), jitter, harmonicity parameters, Pitch Period Entropy (PPE), Detrended Fluctuation Analysis (DFA), etc. [
14,
15,
21,
22,
43]. In [
16], these features were classified as baseline features. Additionally, Praat acoustic analysis software [
44] was used to extract these baseline features. The description of the features is provided in
Table 2. Feature engineering is adopted from [
16] in our study.
The shape of the human vocal tracts (e.g., teeth, tongue etc.) is the most important component of any sound generation. To accurately represent this sound, this shape must be determined correctly. The vocal tract representative is the envelope of the time power spectrum. This envelope is accurately represented by Mel-Frequency Cepstral Coefficients (MFCCs). In other words, MFCCs can imitate the characteristics of the human ear and have been used in different speech recognition tasks [
45,
46]. In this study, MFCCs are being employed to detect the aberration in the human tongue and lips, which are directly affected by PD. In
Figure 1, a stepwise summary of the MFCC block diagram is shown.
The formula for the frequency to Mel scale is given below:
where
f = frequency of the signal.
The Mel-scale relates the acquired frequency of a tone to the actual measured frequency, which scales the frequency to mimic the human ear. Cepstrum is the information rate of change in spectral bands. Mel frequency cepstral is obtained by taking the log of the magnitude of the Fourier spectrum and later taking the spectrum of this log by a cosine transformation; there is a peak observed where there is a periodic element in the original time signal. Upon applying a transform on the frequency spectrum, the resulting spectrum is in neither the time domain nor the frequency domain. Hence, it is called the “quefrency domain” [
47]. The log of the spectrum of the time signal was named cepstrum.
Wavelet transform (WT), similar to Fourier transform, was used with a completely different merit function that uses functions which are confined in both real and Fourier space. The following equation expresses the mathematical function of WT:
where
some function and * is the complex conjugate symbol.
From Equation (2), it can be inferred that WT is an infinite set of various transforms, depending on the merit function used for its computation. It can be useful for making decisions on a signal, especially on the regional scale with small fluctuations. WT is a very popular tool as, in several studies, special features have been extracted from the basic frequency of the speech signal (F0). Speech sample deviation can be captured by WT-based features [
48]. This is how WT can detect sudden aberrations of long-term vowels in clinical speech recordings. In this dataset, the WT-based feature number is 182, which includes the Shannon’s and the log energy entropy, the energy, and the Teager-Kaiser energy of both the approximation and detailed coefficients. WT-based features obtained from the raw (F0) contour and the log transformation of the (F0) contour have been collected using a 10-level discrete wavelet transform.
In a very recent work [
16], TQWT-based features were used. It is a completely discrete, over-complete WT and the main feature extractor [
49]. To transform signals in better quality, TQWT uses three tunable parameters, which are Q-factor (Q), redundancy (r), and the number of levels (J). Speech signals have high oscillatory time series characteristics for which a Q-factor with a relatively high value is appropriate. The TQWT consists of two filter banks. The low pass filter (LPF) output is provided as the inputs of the second LPF or the high pass filter (HPF) bank. The filter banks are iteratively applied. If J is the number of levels, J HPF and one final LPF output will provide J + 1 sub-bands at the end of the decomposition stage. The redundancy rate (r), also known as the decomposition rate, controls the unexpected excessive ringing. Without affecting the shape, this process helps to localize the wavelets in the time domain [
49]. The TQWT parameters are determined in the following order:
Defining the Q-factor parameter to regulate the oscillatory behavior of the wavelet.
Setting the r parameter value greater or equal to three to prevent the undesired ringing in wavelets.
Searching for the best accuracy value in different Q–r pairs of several numbers of levels (J) in the fixed intervals.
There are a total of 432 TQWT-related features available in this data set.
Apart from the aforementioned features, several other features have also been employed depending on vocal fold vibration. Features such as the Vocal Fold Excitation Ratio (VFER), Glottal to Noise Excitation (GNE), the Glottis Quotient (GQ), etc., have also been employed to explore the effect of noise on the vocal fold.
To be mentioned, min-max normalization on this data has been performed to keep the same differences in the range of values by fitting the feature values into a common scale. The normalization process is a data pre-processing part to handle the bias to larger feature values [
50].
5. Results and Analysis
The proposed models were used to detect PD patients. For the experiment, the dataset mentioned in
Section 3 was used to evaluate the model performance to find out the best one, and comparisons with MLP, KNN, RF, and XGBoost were exhibited. The dataset was split into 70% and 30% for training and test purposes, respectively, with no subject occurring in the test or train data simultaneously. In this study, the features stated in
Table 2 were taken as input, and the classification label was given as output. The range of (0–1) was used for the scaling purpose of the original dataset and normalization.
The output of our classifier can be evaluated in terms of accuracy. However, the dataset used in this study had class imbalance with a ratio of 188:64, and so measuring only the accuracy can be misleading in terms of evaluating the classifiers. Therefore, the results of the classifier were measured in terms of the accuracy, F1-score, Mathews Correlation Coefficient (MCC), and Precision-Recall curves. Accuracy can be defined as:
To calculate the F1-score, MCC, and Precision-Recall curve, a confusion matrix is expressed in
Table 3 to understand the binary classifier predictions with
tp,
fp,
tn, and
fn as true positive, false positive, true negative, and false negative, respectively.
From the confusion matrix, the F1-score can be defined as:
In our study, the polynomial kernel function was used with degree 2 for the SVM, the gamma was set to 10 for the non-linear hyperplane, and C was set to 1 to avoid both under-fitting and over-fitting. The best output was obtained from this optimal set of hyper-parameters via k-fold cross-validation, where k was set to 10 for modest variance and low bias with a mean of 0.8 and a standard deviation of 0.06 (approx.). For the first model, including PCA, from
Figure 6, it can be implied that the number of principal components needed to be set to 200 to preserve around 98.8–99% of the total variance of the data and reduce components with insignificant variance from the input.
Table 4 bolsters this intuition of selecting 200 principal components compared with other variable numbers of principal components, fed to the SVM classifier, as it showed the highest accuracy, F1-score, and MCC value of 0.889, 0.928, and 0.7, respectively. Without reducing the number of principal components, and with only the removal of feature redundancy, the training time was 0.203 s, with an accuracy, F1-score, and MCC score of 0.885, 0.925, and 0.688, respectively; whereas with the reduced 200 principal components, the training time was 0.022 s with higher accuracy, F1-score, and MCC score of 0.889, 0.928, and 0.7 respectively.
To evaluate the second model, including the novel deeper SAE with five dense encoding layers with tanh activation and L1 regularization, using adam as the optimizer,
MSE as the loss function, a batch size of 32, and integrated with SVM, the actual raw data points are plotted in
Figure 7 with the two most important features after using t-SNE [
64]. We initialized t-SNE with dimensions 2, perplexity 50, seed 24,680, and random state 0. t-SNE is used to learn, investigate, or evaluate segmentation. It can be employed to visualize the clear data separation.
Figure 7 depicts that the data points were not easily separable, and the features had a non-linear relationship with each other. After applying SAE to the raw data points and feeding the extracted features to the SVM classifier, the best performance was gained by setting the extracted feature number to 180 with comparisons with other feature numbers, as shown in
Table 5.
Figure 8 also shows the data points with the two most important features by using t-SNE, as mentioned earlier, among the extracted 180 features by the SAE. In
Figure 8, the data points are easily separable from the data points in
Figure 7, and it can be seen that the segmentation actually holds up. Juxtaposing
Table 4 and
Table 5, the SAE-SVM model outperformed the former PCA-SVM model with the highest accuracy of 0.935, the highest F1-score of 0.951, and the highest MCC value of 0.788. The non-linear relationship between the vocal features of the PD patients resulted in the better performance of the SAE-SVM model, which outperformed the former PCA-SVM model as PCA features are linearly uncorrelated. Therefore, the deep neural SAE with multiple hidden layers was more successful in compressing the information into a lower-dimensional feature space.
The MLP, XGBoost, KNN, and RF classifiers were implemented to evaluate the effectiveness of the models in this study. The prediction of PD was made with all the models providing the same set of data. From the experimental result, it can be seen that KNN showed the least efficiency among all the models and XGBoost had a better performance than others, but our proposed SAE-SVM model outperformed all the other models in terms of accuracy, F1-score, and MCC.
Table 6 states the accuracy, F1-score, and MCC value of all the models tested and shows that the proposed SAE-SVM model had the highest accuracy of 0.935, the highest F1-score of 0.951, and the highest MCC value of 0.788. It also shows the performance boost of SVM with dimensionality reduction, which in the case of the SAE-SVM was a 9.5% increase in accuracy, 4.9% increase in F1-score, and 32% increase in MCC score compared to the SVM. In addition, it can be inferred that the SAE-SVM had the lowest misdiagnosis rate with 6.5%, and the PCA-SVM had the second lowest misdiagnosis rate with 11.1%.
Figure 9 illustrates the Precision-Recall curve from the experimental results. It depicts that the KNN model had the lowest AUC value with 0.875 and that XGBoost showed a higher AUC of 0.981, but the proposed SAE-SVM model surpassed both models with an AUC of 0.988.
We also compared our best model with other commonly used feature extraction techniques, including the LDA and ICA implementing and incorporating with the same SVM model.
Table 7 shows that our best model of novel deeper SAE with SVM performed better in terms of accuracy, F1-score, and MCC value than the LDA and ICA, as our deeper SAE was more efficient in reducing the dimensionality of feature space and improving the network’s generalization property.
Along with our implemented models, we compared our result with two recent studies made by Sakar et al. [
16], in which an SVM using RBF kernel gave the best result, and Gunduz [
24], in which the CNN framework had the best output. We compared the results of our model with their best results, as they used the same dataset. From
Table 8, it can be seen that our proposed SAE-SVM model outperformed both studies in terms of accuracy, F1-score, and MCC value.
We also used SMOTE for oversampling. From
Table 9, our models showed better accuracy, F1 score, and MCC value in the balanced-class scenario.
6. Discussion
In this study regarding the detection of PD patients from vocal signals, we depicted and implemented two models based on two different feature extraction algorithms along with SVM, which is a popular supervised algorithm in the area of classification problems, using hyperplanes to classify both linear and non-linear dataset. In the first model, a PCA was used as it is a popular unsupervised method for finding the principal components of data in order to reduce the dimensions. This, in turn, bypassed the disadvantage of SVM with decreasing classification performance while having a higher number of features than the number of samples as in the dataset used in this study. In the second model, a novel deeper SAE was developed and used for the same purpose, which is a DNN and works in an unsupervised manner to map the features to a new feature space, reducing the curse of dimensionality problem.
The above-mentioned models were trained and tested using the dataset obtained from the University of California-Irvine (UCI) Machine Learning repository. Due to the high imbalance in the dataset, the F1-score, MCC, and Precision-Recall curve were used to evaluate the models along with accuracy.
For the evaluation of models, all the feature sets, including baseline features, time-frequency features, MFCCs, Wavelet Transform-based features, vocal fold features, and TQWT, were concatenated for the feature extraction purpose. For the first model, 200 principal components were selected from the explained variance of the dataset. Later these 200 principal components were fed into the SVM classifier with a polynomial kernel of degree 2, and it was compared against other variable numbers of principal components. The highest accuracy, F1-score, and MCC were acquired when the number of principal components was set to 200. For the second model, the novel deeper SAE converted the features into 180 features which also distributed the data points in a more easily separable fashion with the polynomial SVM. Other variable numbers of features were also tested, and the results were not as satisfactory as the 180 extracted features. From both the models, it can be implied that the SAE-SVM showed an increase in accuracy rate by 5% (approx.) than that of the PCA-SVM. This was also seen in the F1-score and MCC rate as they increased from 0.928 to 0.951 and 0.7 to 0.788, respectively. Moreover, according to the present study, the best model (SAE-SVM) outperformed other models, including XGBoost, MLP, KNN, and RF. It also outperformed the SVM with other common feature extraction techniques, LDA and ICA. Moreover, it was successful at surpassing the two most recent works of Sakar et al. [
16] and Gunduz [
24] in terms of the accuracy, F1-score, and MCC metrics, and had the lowest misdiagnosis rate of 6.5%. It can be concluded that our proposed SAE-SVM model is a good alternative to both of the models proposed in these two literatures.
The salient advantages of the proposed best model of this study over the previous PD detection studies are stated below:
Different feature extraction techniques were applied, and the relative comparisons were depicted with a much larger dataset with 752 features and 756 voice samples, unlike the recent study in Hemmerling and Sztahó [
28], which has only 198 voice samples and 33 features. Small datasets have several disadvantages, which lead to lower precision in the prediction, lower power, and pose a larger risk by comparing the classes unfairly, even in the circumstances that the data is from a randomized trial. They used PCA only to remove feature redundancy; in contrast, in this study, PCA was used to reduce dimensionality as well as remove feature redundancy, which boosted the training time efficiency and all the performance metrics of our model for a larger dataset.
In our study, the non-linear relationship between vocal features of the PD patients was depicted in
Figure 7. For these non-linear feature relations, our second model SAE-SVM outperformed the former PCA-SVM model as PCA works well with linear relationships. Therefore, the novel deep neural SAE with multiple hidden layers was more successful in compressing the information better into a lower-dimensional feature space. Deep feature extraction of SAE augmented the discriminatory power of distinguishing PD patients as established by the increased MCC value.
As in other previous literatures, if accuracy was used as the only evaluation metric, it can be misleading in the case of an imbalanced dataset. However, inspired by Sakar et al. [
16], we used F1-score and MCC along with accuracy. We also used the Precision-Recall curve to visualize the performance of the models for such skewed class distribution. We also applied SMOTE to synthesize new minority examples to evaluate the models in a balanced-class scenario. Both models showed better balanced-class performance.
The study explored the field of Parkinson’s disease patient detection based on vocal features by building the idea of merging feature extractions, removal of irrelevant data by reducing dimensionality based on the variance of data and additionally using DNN in an unsupervised manner with SVM, which is one of the most powerful classifiers thus far when it comes to data points separable with a larger number of hyperplanes. Imbalanced data deters from having an accurate picture in the detection of PD patients, which can be solved with a more balanced dataset. Further research and experiments can be conducted by employing other dimensionality reduction and feature extraction algorithms such as kernel PCA (kPCA), Denoising Autoencoders to reduce the noise effects of voice signals, etc. The performance of the model can also be improved by applying enhancement algorithms to reduce reverberation, background noise and non-linear distortion [
68,
69]. Along with these, the performance of the proposed model can be further improved with the inclusion of wearable sensor data for measuring tremors and postural instability of individuals to detect the PD features more accurately.