Off the Clock: From Circadian Disruption to Metabolic Disease
Abstract
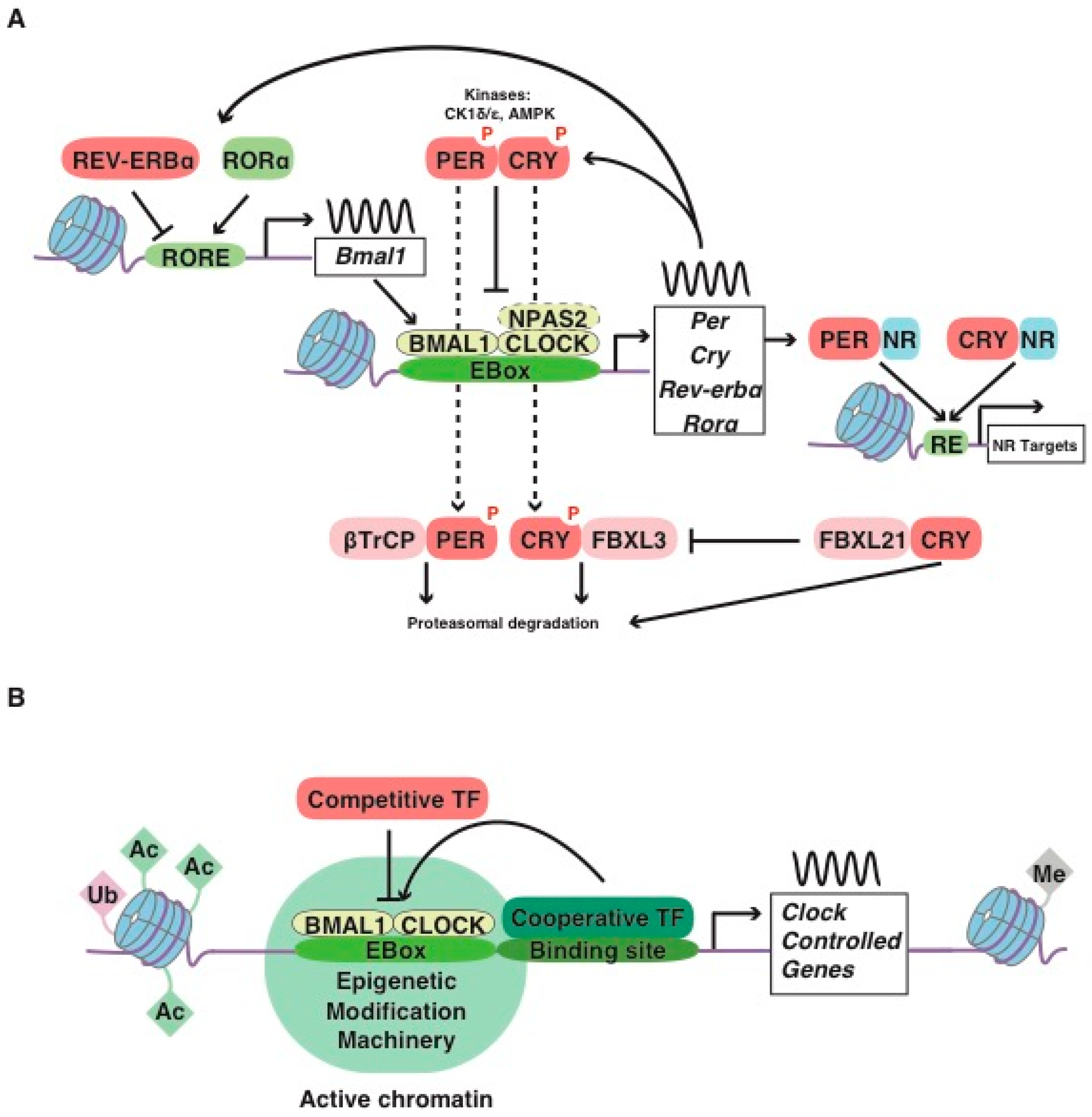
:1. The Mammalian Molecular Clockwork: A Short Story
2. Neural Basis of Mammalian Circadian Behavior and Its Interconnection with Sleep and Energy Centers
3. Clock Genes, Sleep and Metabolism
4. The Emerging Field of Chrono-Pharmacology
5. Conclusions
Funding
Conflicts of Interest
References
- Bell-Pedersen, D.; Cassone, V.M.; Earnest, D.J.; Golden, S.S.; Hardin, P.E.; Thomas, T.L.; Zoran, M.J. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat. Rev. Genet. 2005, 6, 544–556. [Google Scholar] [CrossRef] [PubMed]
- Chiou, Y.Y.; Yang, Y.; Rashid, N.; Ye, R.; Selby, C.P.; Sancar, A. Mammalian Period represses and de-represses transcription by displacing CLOCK-BMAL1 from promoters in a Cryptochrome-dependent manner. Proc. Natl. Acad. Sci. USA 2016, 113, E6072–E6079. [Google Scholar] [CrossRef] [PubMed]
- Ye, R.; Selby, C.P.; Chiou, Y.Y.; Ozkan-Dagliyan, I.; Gaddameedhi, S.; Sancar, A. Dual modes of CLOCK:BMAL1 inhibition mediated by Cryptochrome and Period proteins in the mammalian circadian clock. Genes Dev. 2014, 28, 1989–1998. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Gustafson, C.L.; Sammons, P.J.; Khan, S.K.; Parsley, N.C.; Ramanathan, C.; Lee, H.W.; Liu, A.C.; Partch, C.L. Cryptochrome 1 regulates the circadian clock through dynamic interactions with the BMAL1 C terminus. Nat. Struct. Mol. Biol. 2015, 22, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017, 18, 164–179. [Google Scholar] [CrossRef]
- Grimaldi, B.; Bellet, M.M.; Katada, S.; Astarita, G.; Hirayama, J.; Amin, R.H.; Granneman, J.G.; Piomelli, D.; Leff, T.; Sassone-Corsi, P. PER2 controls lipid metabolism by direct regulation of PPARgamma. Cell Metab. 2010, 12, 509–520. [Google Scholar] [CrossRef]
- Han, D.H.; Lee, Y.J.; Kim, K.; Kim, C.J.; Cho, S. Modulation of glucocorticoid receptor induction properties by core circadian clock proteins. Mol. Cell. Endocrinol. 2014, 383, 170–180. [Google Scholar] [CrossRef]
- Kriebs, A.; Jordan, S.D.; Soto, E.; Henriksson, E.; Sandate, C.R.; Vaughan, M.E.; Chan, A.B.; Duglan, D.; Papp, S.J.; Huber, A.L.; et al. Circadian repressors CRY1 and CRY2 broadly interact with nuclear receptors and modulate transcriptional activity. Proc. Natl. Acad. Sci. USA 2017, 114, 8776–8781. [Google Scholar] [CrossRef]
- Schmutz, I.; Ripperger, J.A.; Baeriswyl-Aebischer, S.; Albrecht, U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010, 24, 345–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asher, G.; Gatfield, D.; Stratmann, M.; Reinke, H.; Dibner, C.; Kreppel, F.; Mostoslavsky, R.; Alt, F.W.; Schibler, U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 2008, 134, 317–328. [Google Scholar] [CrossRef]
- Nakahata, Y.; Kaluzova, M.; Grimaldi, B.; Sahar, S.; Hirayama, J.; Chen, D.; Guarente, L.P.; Sassone-Corsi, P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 2008, 134, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Curtis, A.M.; Seo, S.B.; Westgate, E.J.; Rudic, R.D.; Smyth, E.M.; Chakravarti, D.; FitzGerald, G.A.; McNamara, P. Histone acetyltransferase-dependent chromatin remodeling and the vascular clock. J. Biol. Chem. 2004, 279, 7091–7097. [Google Scholar] [CrossRef] [PubMed]
- Etchegaray, J.P.; Lee, C.; Wade, P.A.; Reppert, S.M. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 2003, 421, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Masri, S.; Rigor, P.; Cervantes, M.; Ceglia, N.; Sebastian, C.; Xiao, C.; Roqueta-Rivera, M.; Deng, C.; Osborne, T.F.; Mostoslavsky, R.; et al. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell 2014, 158, 659–672. [Google Scholar] [CrossRef] [PubMed]
- DiTacchio, L.; Le, H.D.; Vollmers, C.; Hatori, M.; Witcher, M.; Secombe, J.; Panda, S. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science 2011, 333, 1881–1885. [Google Scholar] [CrossRef] [PubMed]
- Valekunja, U.K.; Edgar, R.S.; Oklejewicz, M.; van der Horst, G.T.; O’Neill, J.S.; Tamanini, F.; Turner, D.J.; Reddy, A.B. Histone methyltransferase MLL3 contributes to genome-scale circadian transcription. Proc. Natl. Acad. Sci. USA 2013, 110, 1554–1559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimomura, K.; Kumar, V.; Koike, N.; Kim, T.K.; Chong, J.; Buhr, E.D.; Whiteley, A.R.; Low, S.S.; Omura, C.; Fenner, D.; et al. Usf1, a suppressor of the circadian Clock mutant, reveals the nature of the DNA-binding of the CLOCK:BMAL1 complex in mice. Elife 2013, 2, e00426. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.K.; Maury, E.; Ramsey, K.M.; Perelis, M.; Marcheva, B.; Omura, C.; Kobayashi, Y.; Guttridge, D.C.; Barish, G.D.; Bass, J. Requirement for NF-kappaB in maintenance of molecular and behavioral circadian rhythms in mice. Genes Dev. 2018, 32, 1367–1379. [Google Scholar] [CrossRef] [PubMed]
- Qu, M.; Duffy, T.; Hirota, T.; Kay, S.A. Nuclear receptor HNF4A transrepresses CLOCK:BMAL1 and modulates tissue-specific circadian networks. Proc. Natl. Acad. Sci. USA 2018, 115, E12305–E12312. [Google Scholar] [CrossRef]
- Altman, B.J.; Hsieh, A.L.; Sengupta, A.; Krishnanaiah, S.Y.; Stine, Z.E.; Walton, Z.E.; Gouw, A.M.; Venkataraman, A.; Li, B.; Goraksha-Hicks, P.; et al. MYC Disrupts the Circadian Clock and Metabolism in Cancer Cells. Cell Metab. 2015, 22, 1009–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Fang, B.; Emmett, M.J.; Damle, M.; Sun, Z.; Feng, D.; Armour, S.M.; Remsberg, J.R.; Jager, J.; Soccio, R.E.; et al. GENE REGULATION. Discrete functions of nuclear receptor Rev-erbalpha couple metabolism to the clock. Science 2015, 348, 1488–1492. [Google Scholar] [CrossRef]
- Perelis, M.; Marcheva, B.; Ramsey, K.M.; Schipma, M.J.; Hutchison, A.L.; Taguchi, A.; Peek, C.B.; Hong, H.; Huang, W.; Omura, C.; et al. Pancreatic beta cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science 2015, 350, aac4250. [Google Scholar] [CrossRef]
- Wu, Y.; Tang, D.; Liu, N.; Xiong, W.; Huang, H.; Li, Y.; Ma, Z.; Zhao, H.; Chen, P.; Qi, X.; et al. Reciprocal Regulation between the Circadian Clock and Hypoxia Signaling at the Genome Level in Mammals. Cell Metab. 2017, 25, 73–85. [Google Scholar] [CrossRef]
- Cho, H.; Zhao, X.; Hatori, M.; Yu, R.T.; Barish, G.D.; Lam, M.T.; Chong, L.W.; DiTacchio, L.; Atkins, A.R.; Glass, C.K.; et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature 2012, 485, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Bugge, A.; Feng, D.; Everett, L.J.; Briggs, E.R.; Mullican, S.E.; Wang, F.; Jager, J.; Lazar, M.A. Rev-erbalpha and Rev-erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012, 26, 657–667. [Google Scholar] [CrossRef]
- Shalev, M.; Aviram, R.; Adamovich, Y.; Kraut-Cohen, J.; Shamia, T.; Ben-Dor, S.; Golik, M.; Asher, G. The PXDLS linear motif regulates circadian rhythmicity through protein-protein interactions. Nucleic Acids Res. 2014, 42, 11879–11890. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Lahens, N.F.; Ballance, H.I.; Hughes, M.E.; Hogenesch, J.B. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. USA 2014, 111, 16219–16224. [Google Scholar] [CrossRef] [Green Version]
- Lamia, K.A.; Papp, S.J.; Yu, R.T.; Barish, G.D.; Uhlenhaut, N.H.; Jonker, J.W.; Downes, M.; Evans, R.M. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 2011, 480, 552–556. [Google Scholar] [CrossRef] [Green Version]
- Menet, J.S.; Pescatore, S.; Rosbash, M. CLOCK:BMAL1 is a pioneer-like transcription factor. Genes Dev. 2014, 28, 8–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, A.J.; Menet, J.S. Regulation of circadian clock transcriptional output by CLOCK:BMAL1. PLoS Genet. 2018, 14, e1007156. [Google Scholar] [CrossRef]
- Koike, N.; Yoo, S.H.; Huang, H.C.; Kumar, V.; Lee, C.; Kim, T.K.; Takahashi, J.S. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 2012, 338, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Nakahata, Y.; Sahar, S.; Astarita, G.; Kaluzova, M.; Sassone-Corsi, P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 2009, 324, 654–657. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, K.M.; Yoshino, J.; Brace, C.S.; Abrassart, D.; Kobayashi, Y.; Marcheva, B.; Hong, H.K.; Chong, J.L.; Buhr, E.D.; Lee, C.; et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 2009, 324, 651–654. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Arnal, L.; Hakim, O.; Patel, V.R.; Baldi, P.; Hager, G.L.; Sassone-Corsi, P. Cycles in spatial and temporal chromosomal organization driven by the circadian clock. Nat. Struct. Mol. Biol. 2013, 20, 1206–1213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.H.; Marhon, S.A.; Zhang, Y.; Steger, D.J.; Won, K.J.; Lazar, M.A. Rev-erbalpha dynamically modulates chromatin looping to control circadian gene transcription. Science 2018, 359, 1274–1277. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Liu, T.; Sun, Z.; Bugge, A.; Mullican, S.E.; Alenghat, T.; Liu, X.S.; Lazar, M.A. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 2011, 331, 1315–1319. [Google Scholar] [CrossRef] [PubMed]
- Gu, B.; Swigut, T.; Spencley, A.; Bauer, M.R.; Chung, M.; Meyer, T.; Wysocka, J. Transcription-coupled changes in nuclear mobility of mammalian cis-regulatory elements. Science 2018, 359, 1050–1055. [Google Scholar] [CrossRef]
- Beytebiere, J.R.; Trott, A.J.; Greenwell, B.J.; Osborne, C.A.; Vitet, H.; Spence, J.; Yoo, S.H.; Chen, Z.; Takahashi, J.S.; Ghaffari, N.; et al. Tissue-specific BMAL1 cistromes reveal that rhythmic transcription is associated with rhythmic enhancer-enhancer interactions. Genes Dev. 2019. [Google Scholar] [CrossRef] [PubMed]
- Heinz, S.; Texari, L.; Hayes, M.G.B.; Urbanowski, M.; Chang, M.W.; Givarkes, N.; Rialdi, A.; White, K.M.; Albrecht, R.A.; Pache, L.; et al. Transcription Elongation Can Affect Genome 3D Structure. Cell 2018, 174, 1522–1536. [Google Scholar] [CrossRef] [PubMed]
- Fang, B.; Everett, L.J.; Jager, J.; Briggs, E.; Armour, S.M.; Feng, D.; Roy, A.; Gerhart-Hines, Z.; Sun, Z.; Lazar, M.A. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell 2014, 159, 1140–1152. [Google Scholar] [CrossRef]
- Wang, Y.; Kuang, Z.; Yu, X.; Ruhn, K.A.; Kubo, M.; Hooper, L.V. The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science 2017, 357, 912–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gachon, F.; Olela, F.F.; Schaad, O.; Descombes, P.; Schibler, U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006, 4, 25–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rey, G.; Cesbron, F.; Rougemont, J.; Reinke, H.; Brunner, M.; Naef, F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011, 9, e1000595. [Google Scholar] [CrossRef]
- Gachon, F.; Fonjallaz, P.; Damiola, F.; Gos, P.; Kodama, T.; Zakany, J.; Duboule, D.; Petit, B.; Tafti, M.; Schibler, U. The loss of circadian PAR bZip transcription factors results in epilepsy. Genes Dev. 2004, 18, 1397–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paranjpe, D.A.; Sharma, V.K. Evolution of temporal order in living organisms. J. Circadian Rhythms 2005, 3, 7. [Google Scholar] [CrossRef]
- Pittendrigh, C.S. Temporal organization: Reflections of a Darwinian clock-watcher. Annu. Rev. Physiol. 1993, 55, 16–54. [Google Scholar] [CrossRef] [PubMed]
- Mauvoisin, D.; Wang, J.; Jouffe, C.; Martin, E.; Atger, F.; Waridel, P.; Quadroni, M.; Gachon, F.; Naef, F. Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc. Natl. Acad. Sci. USA 2014, 111, 167–172. [Google Scholar] [CrossRef]
- Krishnaiah, S.Y.; Wu, G.; Altman, B.J.; Growe, J.; Rhoades, S.D.; Coldren, F.; Venkataraman, A.; Olarerin-George, A.O.; Francey, L.J.; Mukherjee, S.; et al. Clock Regulation of Metabolites Reveals Coupling between Transcription and Metabolism. Cell Metab. 2017, 25, 961–974. [Google Scholar] [CrossRef] [PubMed]
- Mure, L.S.; Le, H.D.; Benegiamo, G.; Chang, M.W.; Rios, L.; Jillani, N.; Ngotho, M.; Kariuki, T.; Dkhissi-Benyahya, O.; Cooper, H.M.; et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 2018, 359. [Google Scholar] [CrossRef]
- Herzog, E.D. Neurons and networks in daily rhythms. Nat. Rev. Neurosci. 2007, 8, 790–802. [Google Scholar] [CrossRef]
- Mohawk, J.A.; Green, C.B.; Takahashi, J.S. Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 2012, 35, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Welsh, D.K.; Takahashi, J.S.; Kay, S.A. Suprachiasmatic nucleus: Cell autonomy and network properties. Annu. Rev. Physiol. 2010, 72, 551–577. [Google Scholar] [CrossRef]
- Herzog, E.D.; Hermanstyne, T.; Smyllie, N.J.; Hastings, M.H. Regulating the Suprachiasmatic Nucleus (SCN) Circadian Clockwork: Interplay between Cell-Autonomous and Circuit-Level Mechanisms. Cold Spring Harb. Perspect. Biol. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Rosensweig, C.; Green, C.B. Periodicity, repression, and the molecular architecture of the mammalian circadian clock. Eur. J. Neurosci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Vitaterna, M.H.; King, D.P.; Chang, A.M.; Kornhauser, J.M.; Lowrey, P.L.; McDonald, J.D.; Dove, W.F.; Pinto, L.H.; Turek, F.W.; Takahashi, J.S. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 1994, 264, 719–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, D.P.; Zhao, Y.; Sangoram, A.M.; Wilsbacher, L.D.; Tanaka, M.; Antoch, M.P.; Steeves, T.D.; Vitaterna, M.H.; Kornhauser, J.M.; Lowrey, P.L.; et al. Positional cloning of the mouse circadian clock gene. Cell 1997, 89, 641–653. [Google Scholar] [CrossRef]
- Gekakis, N.; Staknis, D.; Nguyen, H.B.; Davis, F.C.; Wilsbacher, L.D.; King, D.P.; Takahashi, J.S.; Weitz, C.J. Role of the CLOCK protein in the mammalian circadian mechanism. Science 1998, 280, 1564–1569. [Google Scholar] [CrossRef]
- Miller, B.H.; McDearmon, E.L.; Panda, S.; Hayes, K.R.; Zhang, J.; Andrews, J.L.; Antoch, M.P.; Walker, J.R.; Esser, K.A.; Hogenesch, J.B.; et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc. Natl. Acad. Sci. USA 2007, 104, 3342–3347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parsons, M.J.; Brancaccio, M.; Sethi, S.; Maywood, E.S.; Satija, R.; Edwards, J.K.; Jagannath, A.; Couch, Y.; Finelli, M.J.; Smyllie, N.J.; et al. The Regulatory Factor ZFHX3 Modifies Circadian Function in SCN via an AT Motif-Driven Axis. Cell 2015, 162, 607–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedont, J.L.; LeGates, T.A.; Slat, E.A.; Byerly, M.S.; Wang, H.; Hu, J.; Rupp, A.C.; Qian, J.; Wong, G.W.; Herzog, E.D.; et al. Lhx1 controls terminal differentiation and circadian function of the suprachiasmatic nucleus. Cell Rep. 2014, 7, 609–622. [Google Scholar] [CrossRef]
- Chen, S.K.; Badea, T.C.; Hattar, S. Photoentrainment and pupillary light reflex are mediated by distinct populations of ipRGCs. Nature 2011, 476, 92–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golombek, D.A.; Rosenstein, R.E. Physiology of circadian entrainment. Physiol. Rev. 2010, 90, 1063–1102. [Google Scholar] [CrossRef]
- Antle, M.C.; Smith, V.M.; Sterniczuk, R.; Yamakawa, G.R.; Rakai, B.D. Physiological responses of the circadian clock to acute light exposure at night. Rev. Endocr. Metab. Disord. 2009, 10, 279–291. [Google Scholar] [CrossRef]
- Moore, R.Y.; Silver, R. Suprachiasmatic nucleus organization. Chronobiol. Int. 1998, 15, 475–487. [Google Scholar] [CrossRef]
- Yan, L.; Okamura, H. Gradients in the circadian expression of Per1 and Per2 genes in the rat suprachiasmatic nucleus. Eur. J. Neurosci. 2002, 15, 1153–1162. [Google Scholar] [CrossRef]
- Nakamura, W.; Yamazaki, S.; Takasu, N.N.; Mishima, K.; Block, G.D. Differential response of Period 1 expression within the suprachiasmatic nucleus. J. Neurosci. 2005, 25, 5481–5487. [Google Scholar] [CrossRef] [PubMed]
- Hamada, T.; Antle, M.C.; Silver, R. Temporal and spatial expression patterns of canonical clock genes and clock-controlled genes in the suprachiasmatic nucleus. Eur. J. Neurosci. 2004, 19, 1741–1748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, S.; Harang, R.; Meeker, K.; Granados-Fuentes, D.; Tsai, C.A.; Mazuski, C.; Kim, J.; Doyle, F.J., 3rd; Petzold, L.R.; Herzog, E.D. A neuropeptide speeds circadian entrainment by reducing intercellular synchrony. Proc. Natl. Acad. Sci. USA 2013, 110, E4355–E4361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colwell, C.S.; Michel, S.; Itri, J.; Rodriguez, W.; Tam, J.; Lelievre, V.; Hu, Z.; Liu, X.; Waschek, J.A. Disrupted circadian rhythms in VIP- and PHI-deficient mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003, 285, R939–R949. [Google Scholar] [CrossRef]
- Harmar, A.J.; Marston, H.M.; Shen, S.; Spratt, C.; West, K.M.; Sheward, W.J.; Morrison, C.F.; Dorin, J.R.; Piggins, H.D.; Reubi, J.C.; et al. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell 2002, 109, 497–508. [Google Scholar] [CrossRef]
- Mieda, M.; Ono, D.; Hasegawa, E.; Okamoto, H.; Honma, K.; Honma, S.; Sakurai, T. Cellular clocks in AVP neurons of the SCN are critical for interneuronal coupling regulating circadian behavior rhythm. Neuron 2015, 85, 1103–1116. [Google Scholar] [CrossRef]
- Vosko, A.M.; Schroeder, A.; Loh, D.H.; Colwell, C.S. Vasoactive intestinal peptide and the mammalian circadian system. Gen. Comp. Endocrinol. 2007, 152, 165–175. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Suzuki, T.; Mizoro, Y.; Kori, H.; Okada, K.; Chen, Y.; Fustin, J.M.; Yamazaki, F.; Mizuguchi, N.; Zhang, J.; et al. Mice genetically deficient in vasopressin V1a and V1b receptors are resistant to jet lag. Science 2013, 342, 85–90. [Google Scholar] [CrossRef]
- Lee, I.T.; Chang, A.S.; Manandhar, M.; Shan, Y.; Fan, J.; Izumo, M.; Ikeda, Y.; Motoike, T.; Dixon, S.; Seinfeld, J.E.; et al. Neuromedin s-producing neurons act as essential pacemakers in the suprachiasmatic nucleus to couple clock neurons and dictate circadian rhythms. Neuron 2015, 85, 1086–1102. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.R.; Tackenberg, M.C.; McMahon, D.G. Manipulating circadian clock neuron firing rate resets molecular circadian rhythms and behavior. Nat. Neurosci. 2015, 18, 373–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grippo, R.M.; Purohit, A.M.; Zhang, Q.; Zweifel, L.S.; Guler, A.D. Direct Midbrain Dopamine Input to the Suprachiasmatic Nucleus Accelerates Circadian Entrainment. Curr. Biol. 2017, 27, 2465–2475. [Google Scholar] [CrossRef] [PubMed]
- Colwell, C.S. Linking neural activity and molecular oscillations in the SCN. Nat. Rev. Neurosci. 2011, 12, 553–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, G.; Nitabach, M.N. Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J. Neurosci. 2008, 28, 6493–6501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albus, H.; Bonnefont, X.; Chaves, I.; Yasui, A.; Doczy, J.; van der Horst, G.T.; Meijer, J.H. Cryptochrome-deficient mice lack circadian electrical activity in the suprachiasmatic nuclei. Curr. Biol. 2002, 12, 1130–1133. [Google Scholar] [CrossRef]
- Michel, S.; Geusz, M.E.; Zaritsky, J.J.; Block, G.D. Circadian rhythm in membrane conductance expressed in isolated neurons. Science 1993, 259, 239–241. [Google Scholar] [CrossRef] [Green Version]
- Welsh, D.K.; Logothetis, D.E.; Meister, M.; Reppert, S.M. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 1995, 14, 697–706. [Google Scholar] [CrossRef] [Green Version]
- de Jeu, M.; Hermes, M.; Pennartz, C. Circadian modulation of membrane properties in slices of rat suprachiasmatic nucleus. Neuroreport 1998, 9, 3725–3729. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.N.; Nitabach, M.N.; Colwell, C.S. Membrane Currents, Gene Expression, and Circadian Clocks. Cold Spring Harb. Perspect. Biol. 2017, 9. [Google Scholar] [CrossRef]
- Flourakis, M.; Kula-Eversole, E.; Hutchison, A.L.; Han, T.H.; Aranda, K.; Moose, D.L.; White, K.P.; Dinner, A.R.; Lear, B.C.; Ren, D.; et al. A Conserved Bicycle Model for Circadian Clock Control of Membrane Excitability. Cell 2015, 162, 836–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brancaccio, M.; Edwards, M.D.; Patton, A.P.; Smyllie, N.J.; Chesham, J.E.; Maywood, E.S.; Hastings, M.H. Cell-autonomous clock of astrocytes drives circadian behavior in mammals. Science 2019, 363, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Brancaccio, M.; Patton, A.P.; Chesham, J.E.; Maywood, E.S.; Hastings, M.H. Astrocytes Control Circadian Timekeeping in the Suprachiasmatic Nucleus via Glutamatergic Signaling. Neuron 2017, 93, 1420–1435. [Google Scholar] [CrossRef]
- Saper, C.B. The central circadian timing system. Curr. Opin. Neurobiol. 2013, 23, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Reutrakul, S.; Van Cauter, E. Sleep influences on obesity, insulin resistance, and risk of type 2 diabetes. Metabolism 2018, 84, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Vanitallie, T.B. Sleep and energy balance: Interactive homeostatic systems. Metabolism 2006, 55, S30–S35. [Google Scholar] [CrossRef]
- Gizowski, C.; Zaelzer, C.; Bourque, C.W. Clock-driven vasopressin neurotransmission mediates anticipatory thirst prior to sleep. Nature 2016, 537, 685–688. [Google Scholar] [CrossRef]
- Schmidt, C.; Collette, F.; Leclercq, Y.; Sterpenich, V.; Vandewalle, G.; Berthomier, P.; Berthomier, C.; Phillips, C.; Tinguely, G.; Darsaud, A.; et al. Homeostatic sleep pressure and responses to sustained attention in the suprachiasmatic area. Science 2009, 324, 516–519. [Google Scholar] [CrossRef]
- Yu, Y.Q.; Barry, D.M.; Hao, Y.; Liu, X.T.; Chen, Z.F. Molecular and neural basis of contagious itch behavior in mice. Science 2017, 355, 1072–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landgraf, D.; Shostak, A.; Oster, H. Clock genes and sleep. Pflugers Arch. 2012, 463, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Toh, K.L.; Jones, C.R.; He, Y.; Eide, E.J.; Hinz, W.A.; Virshup, D.M.; Ptacek, L.J.; Fu, Y.H. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 2001, 291, 1040–1043. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Padiath, Q.S.; Shapiro, R.E.; Jones, C.R.; Wu, S.C.; Saigoh, N.; Saigoh, K.; Ptacek, L.J.; Fu, Y.H. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature 2005, 434, 640–644. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Toh, K.L.; Jones, C.R.; Shin, J.Y.; Fu, Y.H.; Ptacek, L.J. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell 2007, 128, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Hirano, A.; Shi, G.; Jones, C.R.; Lipzen, A.; Pennacchio, L.A.; Xu, Y.; Hallows, W.C.; McMahon, T.; Yamazaki, M.; Ptacek, L.J.; et al. A Cryptochrome 2 mutation yields advanced sleep phase in humans. Elife 2016, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carpen, J.D.; von Schantz, M.; Smits, M.; Skene, D.J.; Archer, S.N. A silent polymorphism in the PER1 gene associates with extreme diurnal preference in humans. J. Hum. Genet. 2006, 51, 1122–1125. [Google Scholar] [CrossRef] [Green Version]
- Patke, A.; Murphy, P.J.; Onat, O.E.; Krieger, A.C.; Ozcelik, T.; Campbell, S.S.; Young, M.W. Mutation of the Human Circadian Clock Gene CRY1 in Familial Delayed Sleep Phase Disorder. Cell 2017, 169, 203–215. [Google Scholar] [CrossRef]
- Archer, S.N.; Carpen, J.D.; Gibson, M.; Lim, G.H.; Johnston, J.D.; Skene, D.J.; von Schantz, M. Polymorphism in the PER3 promoter associates with diurnal preference and delayed sleep phase disorder. Sleep 2010, 33, 695–701. [Google Scholar] [CrossRef] [PubMed]
- Archer, S.N.; Robilliard, D.L.; Skene, D.J.; Smits, M.; Williams, A.; Arendt, J.; von Schantz, M. A length polymorphism in the circadian clock gene Per3 is linked to delayed sleep phase syndrome and extreme diurnal preference. Sleep 2003, 26, 413–415. [Google Scholar] [CrossRef]
- Ebisawa, T.; Uchiyama, M.; Kajimura, N.; Mishima, K.; Kamei, Y.; Katoh, M.; Watanabe, T.; Sekimoto, M.; Shibui, K.; Kim, K.; et al. Association of structural polymorphisms in the human period3 gene with delayed sleep phase syndrome. EMBO Rep. 2001, 2, 342–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hida, A.; Kitamura, S.; Katayose, Y.; Kato, M.; Ono, H.; Kadotani, H.; Uchiyama, M.; Ebisawa, T.; Inoue, Y.; Kamei, Y.; et al. Screening of clock gene polymorphisms demonstrates association of a PER3 polymorphism with morningness-eveningness preference and circadian rhythm sleep disorder. Sci. Rep. 2014, 4, 6309. [Google Scholar] [CrossRef]
- Turco, M.; Biscontin, A.; Corrias, M.; Caccin, L.; Bano, M.; Chiaromanni, F.; Salamanca, M.; Mattei, D.; Salvoro, C.; Mazzotta, G.; et al. Diurnal preference, mood and the response to morning light in relation to polymorphisms in the human clock gene PER3. Sci. Rep. 2017, 7, 6967. [Google Scholar] [CrossRef]
- Honma, S.; Kawamoto, T.; Takagi, Y.; Fujimoto, K.; Sato, F.; Noshiro, M.; Kato, Y.; Honma, K. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature 2002, 419, 841–844. [Google Scholar] [CrossRef]
- He, Y.; Jones, C.R.; Fujiki, N.; Xu, Y.; Guo, B.; Holder, J.L., Jr.; Rossner, M.J.; Nishino, S.; Fu, Y.H. The transcriptional repressor DEC2 regulates sleep length in mammals. Science 2009, 325, 866–870. [Google Scholar] [CrossRef] [PubMed]
- Balsalobre, A.; Damiola, F.; Schibler, U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 1998, 93, 929–937. [Google Scholar] [CrossRef]
- Stokkan, K.A.; Yamazaki, S.; Tei, H.; Sakaki, Y.; Menaker, M. Entrainment of the circadian clock in the liver by feeding. Science 2001, 291, 490–493. [Google Scholar] [CrossRef] [PubMed]
- Vosko, A.M.; Colwell, C.S.; Avidan, A.Y. Jet lag syndrome: Circadian organization, pathophysiology, and management strategies. Nat. Sci. Sleep 2010, 2, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Turek, F.W.; Joshu, C.; Kohsaka, A.; Lin, E.; Ivanova, G.; McDearmon, E.; Laposky, A.; Losee-Olson, S.; Easton, A.; Jensen, D.R.; et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 2005, 308, 1043–1045. [Google Scholar] [CrossRef] [PubMed]
- Marcheva, B.; Ramsey, K.M.; Buhr, E.D.; Kobayashi, Y.; Su, H.; Ko, C.H.; Ivanova, G.; Omura, C.; Mo, S.; Vitaterna, M.H.; et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 2010, 466, 627–631. [Google Scholar] [CrossRef] [PubMed]
- Barclay, J.L.; Shostak, A.; Leliavski, A.; Tsang, A.H.; Johren, O.; Muller-Fielitz, H.; Landgraf, D.; Naujokat, N.; van der Horst, G.T.; Oster, H. High-fat diet-induced hyperinsulinemia and tissue-specific insulin resistance in Cry-deficient mice. Am. J. Physiol. Endocrinol. Metab. 2013, 304, E1053–E1063. [Google Scholar] [CrossRef] [PubMed]
- Lamia, K.A.; Storch, K.F.; Weitz, C.J. Physiological significance of a peripheral tissue circadian clock. Proc. Natl. Acad. Sci. USA 2008, 105, 15172–15177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, S.Q.; Ansari, T.S.; McGuinness, O.P.; Wasserman, D.H.; Johnson, C.H. Circadian disruption leads to insulin resistance and obesity. Curr. Biol. 2013, 23, 372–381. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Liu, A.; Weidenhammer, A.; Cooksey, R.C.; McClain, D.; Kim, M.K.; Aguilera, G.; Abel, E.D.; Chung, J.H. The role of mPer2 clock gene in glucocorticoid and feeding rhythms. Endocrinology 2009, 150, 2153–2160. [Google Scholar] [CrossRef]
- Paschos, G.K.; Ibrahim, S.; Song, W.L.; Kunieda, T.; Grant, G.; Reyes, T.M.; Bradfield, C.A.; Vaughan, C.H.; Eiden, M.; Masoodi, M.; et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat. Med. 2012, 18, 1768–1777. [Google Scholar] [CrossRef] [Green Version]
- Kettner, N.M.; Mayo, S.A.; Hua, J.; Lee, C.; Moore, D.D.; Fu, L. Circadian Dysfunction Induces Leptin Resistance in Mice. Cell Metab. 2015, 22, 448–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobi, D.; Liu, S.; Burkewitz, K.; Kory, N.; Knudsen, N.H.; Alexander, R.K.; Unluturk, U.; Li, X.; Kong, X.; Hyde, A.L.; et al. Hepatic Bmal1 Regulates Rhythmic Mitochondrial Dynamics and Promotes Metabolic Fitness. Cell Metab. 2015, 22, 709–720. [Google Scholar] [CrossRef]
- Pan, X.; Bradfield, C.A.; Hussain, M.M. Global and hepatocyte-specific ablation of Bmal1 induces hyperlipidaemia and enhances atherosclerosis. Nat. Commun. 2016, 7, 13011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehlen, J.C.; Brager, A.J.; Baggs, J.; Pinckney, L.; Gray, C.L.; DeBruyne, J.P.; Esser, K.A.; Takahashi, J.S.; Paul, K.N. Bmal1 function in skeletal muscle regulates sleep. Elife 2017, 6. [Google Scholar] [CrossRef] [Green Version]
- Dyar, K.A.; Ciciliot, S.; Wright, L.E.; Bienso, R.S.; Tagliazucchi, G.M.; Patel, V.R.; Forcato, M.; Paz, M.I.; Gudiksen, A.; Solagna, F.; et al. Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol. Metab. 2014, 3, 29–41. [Google Scholar] [CrossRef]
- Xie, Z.; Su, W.; Liu, S.; Zhao, G.; Esser, K.; Schroder, E.A.; Lefta, M.; Stauss, H.M.; Guo, Z.; Gong, M.C. Smooth-muscle BMAL1 participates in blood pressure circadian rhythm regulation. J. Clin. Investig. 2015, 125, 324–336. [Google Scholar] [CrossRef]
- Winter, C.; Silvestre-Roig, C.; Ortega-Gomez, A.; Lemnitzer, P.; Poelman, H.; Schumski, A.; Winter, J.; Drechsler, M.; de Jong, R.; Immler, R.; et al. Chrono-pharmacological Targeting of the CCL2-CCR2 Axis Ameliorates Atherosclerosis. Cell Metab. 2018, 28, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Early, J.O.; Menon, D.; Wyse, C.A.; Cervantes-Silva, M.P.; Zaslona, Z.; Carroll, R.G.; Palsson-McDermott, E.M.; Angiari, S.; Ryan, D.G.; Corcoran, S.E.; et al. Circadian clock protein BMAL1 regulates IL-1beta in macrophages via NRF2. Proc. Natl. Acad. Sci. USA 2018, 115, E8460–E8468. [Google Scholar] [CrossRef]
- Cedernaes, J.; Huang, W.; Ramsey, K.M.; Waldeck, N.; Cheng, L.; Marcheva, B.; Omura, C.; Kobayashi, Y.; Peek, C.B.; Levine, D.C.; et al. Transcriptional Basis for Rhythmic Control of Hunger and Metabolism within the AgRP Neuron. Cell Metab. 2019. [Google Scholar] [CrossRef]
- Maury, E.; Ramsey, K.M.; Bass, J. Circadian rhythms and metabolic syndrome: From experimental genetics to human disease. Circ. Res. 2010, 106, 447–462. [Google Scholar] [CrossRef]
- Corella, D.; Asensio, E.M.; Coltell, O.; Sorli, J.V.; Estruch, R.; Martinez-Gonzalez, M.A.; Salas-Salvado, J.; Castaner, O.; Aros, F.; Lapetra, J.; et al. CLOCK gene variation is associated with incidence of type-2 diabetes and cardiovascular diseases in type-2 diabetic subjects: Dietary modulation in the PREDIMED randomized trial. Cardiovasc. Diabetol. 2016, 15, 4. [Google Scholar] [CrossRef] [PubMed]
- Sookoian, S.; Gemma, C.; Gianotti, T.F.; Burgueno, A.; Castano, G.; Pirola, C.J. Genetic variants of Clock transcription factor are associated with individual susceptibility to obesity. Am. J. Clin. Nutr. 2008, 87, 1606–1615. [Google Scholar] [CrossRef] [PubMed]
- Scott, E.M.; Carter, A.M.; Grant, P.J. Association between polymorphisms in the Clock gene, obesity and the metabolic syndrome in man. Int. J. Obes. 2008, 32, 658–662. [Google Scholar] [CrossRef] [PubMed]
- Woon, P.Y.; Kaisaki, P.J.; Braganca, J.; Bihoreau, M.T.; Levy, J.C.; Farrall, M.; Gauguier, D. Aryl hydrocarbon receptor nuclear translocator-like (BMAL1) is associated with susceptibility to hypertension and type 2 diabetes. Proc. Natl. Acad. Sci. USA 2007, 104, 14412–14417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dupuis, J.; Langenberg, C.; Prokopenko, I.; Saxena, R.; Soranzo, N.; Jackson, A.U.; Wheeler, E.; Glazer, N.L.; Bouatia-Naji, N.; Gloyn, A.L.; et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010, 42, 105–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barker, A.; Sharp, S.J.; Timpson, N.J.; Bouatia-Naji, N.; Warrington, N.M.; Kanoni, S.; Beilin, L.J.; Brage, S.; Deloukas, P.; Evans, D.M.; et al. Association of genetic Loci with glucose levels in childhood and adolescence: A meta-analysis of over 6,000 children. Diabetes 2011, 60, 1805–1812. [Google Scholar] [CrossRef]
- Ruano, E.G.; Canivell, S.; Vieira, E. REV-ERB ALPHA polymorphism is associated with obesity in the Spanish obese male population. PLoS ONE 2014, 9, e104065. [Google Scholar] [CrossRef]
- Dashti, H.S.; Follis, J.L.; Smith, C.E.; Tanaka, T.; Garaulet, M.; Gottlieb, D.J.; Hruby, A.; Jacques, P.F.; Kiefte-de Jong, J.C.; Lamon-Fava, S.; et al. Gene-Environment Interactions of Circadian-Related Genes for Cardiometabolic Traits. Diabetes Care 2015, 38, 1456–1466. [Google Scholar] [CrossRef] [Green Version]
- Bonnefond, A.; Clement, N.; Fawcett, K.; Yengo, L.; Vaillant, E.; Guillaume, J.L.; Dechaume, A.; Payne, F.; Roussel, R.; Czernichow, S.; et al. Rare MTNR1B variants impairing melatonin receptor 1B function contribute to type 2 diabetes. Nat. Genet. 2012, 44, 297–301. [Google Scholar] [CrossRef] [Green Version]
- Bouatia-Naji, N.; Bonnefond, A.; Cavalcanti-Proenca, C.; Sparso, T.; Holmkvist, J.; Marchand, M.; Delplanque, J.; Lobbens, S.; Rocheleau, G.; Durand, E.; et al. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat. Genet. 2009, 41, 89–94. [Google Scholar] [CrossRef]
- Lyssenko, V.; Nagorny, C.L.; Erdos, M.R.; Wierup, N.; Jonsson, A.; Spegel, P.; Bugliani, M.; Saxena, R.; Fex, M.; Pulizzi, N.; et al. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat. Genet. 2009, 41, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Prokopenko, I.; Langenberg, C.; Florez, J.C.; Saxena, R.; Soranzo, N.; Thorleifsson, G.; Loos, R.J.; Manning, A.K.; Jackson, A.U.; Aulchenko, Y.; et al. Variants in MTNR1B influence fasting glucose levels. Nat. Genet. 2009, 41, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Tuomi, T.; Nagorny, C.L.F.; Singh, P.; Bennet, H.; Yu, Q.; Alenkvist, I.; Isomaa, B.; Ostman, B.; Soderstrom, J.; Pesonen, A.K.; et al. Increased Melatonin Signaling Is a Risk Factor for Type 2 Diabetes. Cell Metab. 2016, 23, 1067–1077. [Google Scholar] [CrossRef] [PubMed]
- Scheer, F.A.; Hilton, M.F.; Mantzoros, C.S.; Shea, S.A. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc. Natl. Acad. Sci. USA 2009, 106, 4453–4458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammerschlag, A.R.; Stringer, S.; de Leeuw, C.A.; Sniekers, S.; Taskesen, E.; Watanabe, K.; Blanken, T.F.; Dekker, K.; Te Lindert, B.H.W.; Wassing, R.; et al. Genome-wide association analysis of insomnia complaints identifies risk genes and genetic overlap with psychiatric and metabolic traits. Nat. Genet. 2017, 49, 1584–1592. [Google Scholar] [CrossRef] [Green Version]
- Lane, J.M.; Jones, S.E.; Dashti, H.S.; Wood, A.R.; Aragam, K.G.; van Hees, V.T.; Strand, L.B.; Winsvold, B.S.; Wang, H.; Bowden, J.; et al. Biological and clinical insights from genetics of insomnia symptoms. Nat. Genet. 2019. [Google Scholar] [CrossRef] [PubMed]
- Lane, J.M.; Liang, J.; Vlasac, I.; Anderson, S.G.; Bechtold, D.A.; Bowden, J.; Emsley, R.; Gill, S.; Little, M.A.; Luik, A.I.; et al. Genome-wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nat. Genet. 2017, 49, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, F.; Fuster, V.; Fernandez-Alvira, J.M.; Fernandez-Friera, L.; Lopez-Melgar, B.; Blanco-Rojo, R.; Fernandez-Ortiz, A.; Garcia-Pavia, P.; Sanz, J.; Mendiguren, J.M.; et al. Association of Sleep Duration and Quality With Subclinical Atherosclerosis. J. Am. Coll. Cardiol. 2019, 73, 134–144. [Google Scholar] [CrossRef] [PubMed]
- McAlpine, C.S.; Kiss, M.G.; Rattik, S.; He, S.; Vassalli, A.; Valet, C.; Anzai, A.; Chan, C.T.; Mindur, J.E.; Kahles, F.; et al. Sleep modulates haematopoiesis and protects against atherosclerosis. Nature 2019, 566, 383–387. [Google Scholar] [CrossRef]
- Leproult, R.; Holmback, U.; Van Cauter, E. Circadian misalignment augments markers of insulin resistance and inflammation, independently of sleep loss. Diabetes 2014, 63, 1860–1869. [Google Scholar] [CrossRef] [PubMed]
- Morris, C.J.; Purvis, T.E.; Hu, K.; Scheer, F.A. Circadian misalignment increases cardiovascular disease risk factors in humans. Proc. Natl. Acad. Sci. USA 2016, 113, E1402–E1411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wefers, J.; van Moorsel, D.; Hansen, J.; Connell, N.J.; Havekes, B.; Hoeks, J.; van Marken Lichtenbelt, W.D.; Duez, H.; Phielix, E.; Kalsbeek, A.; et al. Circadian misalignment induces fatty acid metabolism gene profiles and compromises insulin sensitivity in human skeletal muscle. Proc. Natl. Acad. Sci. USA 2018, 115, 7789–7794. [Google Scholar] [CrossRef] [PubMed]
- Depner, C.M.; Melanson, E.L.; McHill, A.W.; Wright, K.P., Jr. Mistimed food intake and sleep alters 24-hour time-of-day patterns of the human plasma proteome. Proc. Natl. Acad. Sci. USA 2018, 115, E5390–E5399. [Google Scholar] [CrossRef] [PubMed]
- Lane, J.M.; Vlasac, I.; Anderson, S.G.; Kyle, S.D.; Dixon, W.G.; Bechtold, D.A.; Gill, S.; Little, M.A.; Luik, A.; Loudon, A.; et al. Genome-wide association analysis identifies novel loci for chronotype in 100,420 individuals from the UK Biobank. Nat. Commun. 2016, 7, 10889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, S.E.; Tyrrell, J.; Wood, A.R.; Beaumont, R.N.; Ruth, K.S.; Tuke, M.A.; Yaghootkar, H.; Hu, Y.; Teder-Laving, M.; Hayward, C.; et al. Genome-Wide Association Analyses in 128,266 Individuals Identifies New Morningness and Sleep Duration Loci. PLoS Genet. 2016, 12, e1006125. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.E.; Lane, J.M.; Wood, A.R.; van Hees, V.T.; Tyrrell, J.; Beaumont, R.N.; Jeffries, A.R.; Dashti, H.S.; Hillsdon, M.; Ruth, K.S.; et al. Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat. Commun. 2019, 10, 343. [Google Scholar] [CrossRef]
- Hu, Y.; Shmygelska, A.; Tran, D.; Eriksson, N.; Tung, J.Y.; Hinds, D.A. GWAS of 89,283 individuals identifies genetic variants associated with self-reporting of being a morning person. Nat. Commun. 2016, 7, 10448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thaiss, C.A.; Zeevi, D.; Levy, M.; Zilberman-Schapira, G.; Suez, J.; Tengeler, A.C.; Abramson, L.; Katz, M.N.; Korem, T.; Zmora, N.; et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 2014, 159, 514–529. [Google Scholar] [CrossRef] [PubMed]
- Mukherji, A.; Kobiita, A.; Ye, T.; Chambon, P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell 2013, 153, 812–827. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Levy, M.; Korem, T.; Dohnalova, L.; Shapiro, H.; Jaitin, D.A.; David, E.; Winter, D.R.; Gury-BenAri, M.; Tatirovsky, E.; et al. Microbiota Diurnal Rhythmicity Programs Host Transcriptome Oscillations. Cell 2016, 167, 1495–1510. [Google Scholar] [CrossRef] [PubMed]
- Zmora, N.; Suez, J.; Elinav, E. You are what you eat: Diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 35–56. [Google Scholar] [CrossRef] [PubMed]
- Kohsaka, A.; Laposky, A.D.; Ramsey, K.M.; Estrada, C.; Joshu, C.; Kobayashi, Y.; Turek, F.W.; Bass, J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007, 6, 414–421. [Google Scholar] [CrossRef]
- Damiola, F.; Le Minh, N.; Preitner, N.; Kornmann, B.; Fleury-Olela, F.; Schibler, U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000, 14, 2950–2961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arble, D.M.; Bass, J.; Laposky, A.D.; Vitaterna, M.H.; Turek, F.W. Circadian timing of food intake contributes to weight gain. Obesity 2009, 17, 2100–2102. [Google Scholar] [CrossRef]
- Buhr, E.D.; Yoo, S.H.; Takahashi, J.S. Temperature as a universal resetting cue for mammalian circadian oscillators. Science 2010, 330, 379–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balsalobre, A.; Brown, S.A.; Marcacci, L.; Tronche, F.; Kellendonk, C.; Reichardt, H.M.; Schutz, G.; Schibler, U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 2000, 289, 2344–2347. [Google Scholar] [CrossRef]
- Hatori, M.; Vollmers, C.; Zarrinpar, A.; DiTacchio, L.; Bushong, E.A.; Gill, S.; Leblanc, M.; Chaix, A.; Joens, M.; Fitzpatrick, J.A.; et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012, 15, 848–860. [Google Scholar] [CrossRef] [PubMed]
- Chaix, A.; Zarrinpar, A.; Miu, P.; Panda, S. Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab. 2014, 20, 991–1005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sutton, E.F.; Beyl, R.; Early, K.S.; Cefalu, W.T.; Ravussin, E.; Peterson, C.M. Early Time-Restricted Feeding Improves Insulin Sensitivity, Blood Pressure, and Oxidative Stress Even without Weight Loss in Men with Prediabetes. Cell Metab. 2018, 27, 1212–1221. [Google Scholar] [CrossRef]
- Chaix, A.; Lin, T.; Le, H.D.; Chang, M.W.; Panda, S. Time-Restricted Feeding Prevents Obesity and Metabolic Syndrome in Mice Lacking a Circadian Clock. Cell Metab. 2019, 29, 303–319. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, J.; Drevet, K.; Pevet, P.; Challet, E. Daily meal timing is not necessary for resetting the main circadian clock by calorie restriction. J. Neuroendocrinol. 2008, 20, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, J.; Gourmelen, S.; Dumont, S.; Sage-Ciocca, D.; Pevet, P.; Challet, E. Setting the main circadian clock of a diurnal mammal by hypocaloric feeding. J. Physiol. 2012, 590, 3155–3168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendoza, J.; Graff, C.; Dardente, H.; Pevet, P.; Challet, E. Feeding cues alter clock gene oscillations and photic responses in the suprachiasmatic nuclei of mice exposed to a light/dark cycle. J. Neurosci. 2005, 25, 1514–1522. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Rodriguez, V.A.; de Groot, M.H.M.; Rijo-Ferreira, F.; Green, C.B.; Takahashi, J.S. Mice under Caloric Restriction Self-Impose a Temporal Restriction of Food Intake as Revealed by an Automated Feeder System. Cell Metab. 2017, 26, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Eckel-Mahan, K.L.; Patel, V.R.; de Mateo, S.; Orozco-Solis, R.; Ceglia, N.J.; Sahar, S.; Dilag-Penilla, S.A.; Dyar, K.A.; Baldi, P.; Sassone-Corsi, P. Reprogramming of the circadian clock by nutritional challenge. Cell 2013, 155, 1464–1478. [Google Scholar] [CrossRef]
- Guan, D.; Xiong, Y.; Borck, P.C.; Jang, C.; Doulias, P.T.; Papazyan, R.; Fang, B.; Jiang, C.; Zhang, Y.; Briggs, E.R.; et al. Diet-Induced Circadian Enhancer Remodeling Synchronizes Opposing Hepatic Lipid Metabolic Processes. Cell 2018, 174, 831–842. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, G.; Zhang, H.; Karin, M.; Bai, H.; Cai, D. Hypothalamic IKKbeta/NF-kappaB and ER stress link overnutrition to energy imbalance and obesity. Cell 2008, 135, 61–73. [Google Scholar] [CrossRef]
- Cai, D.; Yuan, M.; Frantz, D.F.; Melendez, P.A.; Hansen, L.; Lee, J.; Shoelson, S.E. Local and systemic insulin resistance resulting from hepatic activation of IKK-beta and NF-kappaB. Nat. Med. 2005, 11, 183–190. [Google Scholar] [CrossRef]
- Arkan, M.C.; Hevener, A.L.; Greten, F.R.; Maeda, S.; Li, Z.W.; Long, J.M.; Wynshaw-Boris, A.; Poli, G.; Olefsky, J.; Karin, M. IKK-beta links inflammation to obesity-induced insulin resistance. Nat. Med. 2005, 11, 191–198. [Google Scholar] [CrossRef]
- Zaret, K.S.; Carroll, J.S. Pioneer transcription factors: Establishing competence for gene expression. Genes Dev. 2011, 25, 2227–2241. [Google Scholar] [CrossRef]
- Iwafuchi-Doi, M.; Zaret, K.S. Pioneer transcription factors in Cell Rep.rogramming. Genes Dev. 2014, 28, 2679–2692. [Google Scholar] [CrossRef]
- Murakami, M.; Tognini, P.; Liu, Y.; Eckel-Mahan, K.L.; Baldi, P.; Sassone-Corsi, P. Gut microbiota directs PPARgamma-driven reprogramming of the liver circadian clock by nutritional challenge. EMBO Rep. 2016, 17, 1292–1303. [Google Scholar] [CrossRef]
- den Besten, G.; Bleeker, A.; Gerding, A.; van Eunen, K.; Havinga, R.; van Dijk, T.H.; Oosterveer, M.H.; Jonker, J.W.; Groen, A.K.; Reijngoud, D.J.; et al. Short-Chain Fatty Acids Protect Against High-Fat Diet-Induced Obesity via a PPARgamma-Dependent Switch From Lipogenesis to Fat Oxidation. Diabetes 2015, 64, 2398–2408. [Google Scholar] [CrossRef]
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C.; et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007, 56, 1761–1772. [Google Scholar] [CrossRef]
- Ribas-Latre, A.; Fekry, B.; Kwok, C.; Baumgartner, C.; Shivshankar, S.; Sun, K.; Chen, Z.; Eckel-Mahan, K. Rosiglitazone reverses high fat diet-induced changes in BMAL1 function in muscle, fat, and liver tissue in mice. Int. J. Obes. 2018. [Google Scholar] [CrossRef] [PubMed]
- Dyar, K.A.; Lutter, D.; Artati, A.; Ceglia, N.J.; Liu, Y.; Armenta, D.; Jastroch, M.; Schneider, S.; de Mateo, S.; Cervantes, M.; et al. Atlas of Circadian Metabolism Reveals System-wide Coordination and Communication between Clocks. Cell 2018, 174, 1571–1585. [Google Scholar] [CrossRef] [PubMed]
- Toda, H.; Williams, J.A.; Gulledge, M.; Sehgal, A. A sleep-inducing gene, nemuri, links sleep and immune function in Drosophila. Science 2019, 363, 509–515. [Google Scholar] [CrossRef]
- Hanada, R.; Leibbrandt, A.; Hanada, T.; Kitaoka, S.; Furuyashiki, T.; Fujihara, H.; Trichereau, J.; Paolino, M.; Qadri, F.; Plehm, R.; et al. Central control of fever and female body temperature by RANKL/RANK. Nature 2009, 462, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, N.; Levine, B.J.; Loy, K.A.; Duke, W.L.; Meyerson, O.S.; Jamnik, A.A.; Carter, M.E. Hypothalamic Neurons that Regulate Feeding Can Influence Sleep/Wake States Based on Homeostatic Need. Curr. Biol. 2018, 28, 3736–3747. [Google Scholar] [CrossRef]
- Hakansson, M.L.; Brown, H.; Ghilardi, N.; Skoda, R.C.; Meister, B. Leptin receptor immunoreactivity in chemically defined target neurons of the hypothalamus. J. Neurosci. 1998, 18, 559–572. [Google Scholar] [CrossRef]
- Laposky, A.D.; Bradley, M.A.; Williams, D.L.; Bass, J.; Turek, F.W. Sleep-wake regulation is altered in leptin-resistant (db/db) genetically obese and diabetic mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008, 295, R2059–R2066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agudelo, L.Z.; Femenia, T.; Orhan, F.; Porsmyr-Palmertz, M.; Goiny, M.; Martinez-Redondo, V.; Correia, J.C.; Izadi, M.; Bhat, M.; Schuppe-Koistinen, I.; et al. Skeletal muscle PGC-1alpha1 modulates kynurenine metabolism and mediates resilience to stress-induced depression. Cell 2014, 159, 33–45. [Google Scholar] [CrossRef]
- Bookout, A.L.; de Groot, M.H.; Owen, B.M.; Lee, S.; Gautron, L.; Lawrence, H.L.; Ding, X.; Elmquist, J.K.; Takahashi, J.S.; Mangelsdorf, D.J.; et al. FGF21 regulates metabolism and circadian behavior by acting on the nervous system. Nat. Med. 2013, 19, 1147–1152. [Google Scholar] [CrossRef] [PubMed]
- Blum, I.D.; Patterson, Z.; Khazall, R.; Lamont, E.W.; Sleeman, M.W.; Horvath, T.L.; Abizaid, A. Reduced anticipatory locomotor responses to scheduled meals in ghrelin receptor deficient mice. Neuroscience 2009, 164, 351–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horvath, T.L.; Abizaid, A.; Dietrich, M.O.; Li, Y.; Takahashi, J.S.; Bass, J. Ghrelin-immunopositive hypothalamic neurons tie the circadian clock and visual system to the lateral hypothalamic arousal center. Mol. Metab. 2012, 1, 79–85. [Google Scholar] [CrossRef]
- Weikel, J.C.; Wichniak, A.; Ising, M.; Brunner, H.; Friess, E.; Held, K.; Mathias, S.; Schmid, D.A.; Uhr, M.; Steiger, A. Ghrelin promotes slow-wave sleep in humans. Am. J. Physiol. Endocrinol. Metab. 2003, 284, E407–E415. [Google Scholar] [CrossRef]
- Yannielli, P.C.; Molyneux, P.C.; Harrington, M.E.; Golombek, D.A. Ghrelin effects on the circadian system of mice. J. Neurosci. 2007, 27, 2890–2895. [Google Scholar] [CrossRef]
- Revollo, J.R.; Korner, A.; Mills, K.F.; Satoh, A.; Wang, T.; Garten, A.; Dasgupta, B.; Sasaki, Y.; Wolberger, C.; Townsend, R.R.; et al. Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell Metab. 2007, 6, 363–375. [Google Scholar] [CrossRef]
- Yoon, M.J.; Yoshida, M.; Johnson, S.; Takikawa, A.; Usui, I.; Tobe, K.; Nakagawa, T.; Yoshino, J.; Imai, S. SIRT1-Mediated eNAMPT Secretion from Adipose Tissue Regulates Hypothalamic NAD+ and Function in Mice. Cell Metab. 2015, 21, 706–717. [Google Scholar] [CrossRef] [PubMed]
- Lourenco, M.V.; Frozza, R.L.; de Freitas, G.B.; Zhang, H.; Kincheski, G.C.; Ribeiro, F.C.; Goncalves, R.A.; Clarke, J.R.; Beckman, D.; Staniszewski, A.; et al. Exercise-linked FNDC5/irisin rescues synaptic plasticity and memory defects in Alzheimer’s models. Nat. Med. 2019, 25, 165–175. [Google Scholar] [CrossRef]
- Wrann, C.D.; White, J.P.; Salogiannnis, J.; Laznik-Bogoslavski, D.; Wu, J.; Ma, D.; Lin, J.D.; Greenberg, M.E.; Spiegelman, B.M. Exercise induces hippocampal BDNF through a PGC-1alpha/FNDC5 pathway. Cell Metab. 2013, 18, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Clasadonte, J.; Scemes, E.; Wang, Z.; Boison, D.; Haydon, P.G. Connexin 43-Mediated Astroglial Metabolic Networks Contribute to the Regulation of the Sleep-Wake Cycle. Neuron 2017, 95, 1365–1380. [Google Scholar] [CrossRef] [PubMed]
- Rutter, J.; Reick, M.; Wu, L.C.; McKnight, S.L. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science 2001, 293, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Dang, F.; Sun, X.; Ma, X.; Wu, R.; Zhang, D.; Chen, Y.; Xu, Q.; Wu, Y.; Liu, Y. Insulin post-transcriptionally modulates Bmal1 protein to affect the hepatic circadian clock. Nat. Commun. 2016, 7, 12696. [Google Scholar] [CrossRef] [Green Version]
- Xu, A.W.; Kaelin, C.B.; Takeda, K.; Akira, S.; Schwartz, M.W.; Barsh, G.S. PI3K integrates the action of insulin and leptin on hypothalamic neurons. J. Clin. Investig. 2005, 115, 951–958. [Google Scholar] [CrossRef] [Green Version]
- Ramadori, G.; Fujikawa, T.; Fukuda, M.; Anderson, J.; Morgan, D.A.; Mostoslavsky, R.; Stuart, R.C.; Perello, M.; Vianna, C.R.; Nillni, E.A.; et al. SIRT1 deacetylase in POMC neurons is required for homeostatic defenses against diet-induced obesity. Cell Metab. 2010, 12, 78–87. [Google Scholar] [CrossRef] [Green Version]
- Aguilar-Arnal, L.; Ranjit, S.; Stringari, C.; Orozco-Solis, R.; Gratton, E.; Sassone-Corsi, P. Spatial dynamics of SIRT1 and the subnuclear distribution of NADH species. Proc. Natl. Acad. Sci. USA 2016, 113, 12715–12720. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.C.; Guarente, L. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell 2013, 153, 1448–1460. [Google Scholar] [CrossRef] [Green Version]
- Asher, G.; Reinke, H.; Altmeyer, M.; Gutierrez-Arcelus, M.; Hottiger, M.O.; Schibler, U. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell 2010, 142, 943–953. [Google Scholar] [CrossRef]
- Lamia, K.A.; Sachdeva, U.M.; DiTacchio, L.; Williams, E.C.; Alvarez, J.G.; Egan, D.F.; Vasquez, D.S.; Juguilon, H.; Panda, S.; Shaw, R.J.; et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 2009, 326, 437–440. [Google Scholar] [CrossRef]
- Chavan, R.; Feillet, C.; Costa, S.S.; Delorme, J.E.; Okabe, T.; Ripperger, J.A.; Albrecht, U. Liver-derived ketone bodies are necessary for food anticipation. Nat. Commun. 2016, 7, 10580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.K.; Ahima, R.S. Physiology of leptin: Energy homeostasis, neuroendocrine function and metabolism. Metabolism 2015, 64, 24–34. [Google Scholar] [CrossRef]
- Laposky, A.D.; Shelton, J.; Bass, J.; Dugovic, C.; Perrino, N.; Turek, F.W. Altered sleep regulation in leptin-deficient mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006, 290, R894–R903. [Google Scholar] [CrossRef]
- Funato, H.; Tsai, A.L.; Willie, J.T.; Kisanuki, Y.; Williams, S.C.; Sakurai, T.; Yanagisawa, M. Enhanced orexin receptor-2 signaling prevents diet-induced obesity and improves leptin sensitivity. Cell Metab. 2009, 9, 64–76. [Google Scholar] [CrossRef]
- Bostrom, P.; Wu, J.; Jedrychowski, M.P.; Korde, A.; Ye, L.; Lo, J.C.; Rasbach, K.A.; Bostrom, E.A.; Choi, J.H.; Long, J.Z.; et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 2012, 481, 463–468. [Google Scholar] [CrossRef]
- Natalicchio, A.; Marrano, N.; Biondi, G.; Spagnuolo, R.; Labarbuta, R.; Porreca, I.; Cignarelli, A.; Bugliani, M.; Marchetti, P.; Perrini, S.; et al. The Myokine Irisin Is Released in Response to Saturated Fatty Acids and Promotes Pancreatic beta-Cell Survival and Insulin Secretion. Diabetes 2017, 66, 2849–2856. [Google Scholar] [CrossRef]
- Hui, S.; Ghergurovich, J.M.; Morscher, R.J.; Jang, C.; Teng, X.; Lu, W.; Esparza, L.A.; Reya, T.; Le, Z.; Yanxiang Guo, J.; et al. Glucose feeds the TCA cycle via circulating lactate. Nature 2017, 551, 115–118. [Google Scholar] [CrossRef] [Green Version]
- Morland, C.; Andersson, K.A.; Haugen, O.P.; Hadzic, A.; Kleppa, L.; Gille, A.; Rinholm, J.E.; Palibrk, V.; Diget, E.H.; Kennedy, L.H.; et al. Exercise induces cerebral VEGF and angiogenesis via the lactate receptor HCAR1. Nat. Commun. 2017, 8, 15557. [Google Scholar] [CrossRef] [Green Version]
- Kaasik, K.; Kivimae, S.; Allen, J.J.; Chalkley, R.J.; Huang, Y.; Baer, K.; Kissel, H.; Burlingame, A.L.; Shokat, K.M.; Ptacek, L.J.; et al. Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab. 2013, 17, 291–302. [Google Scholar] [CrossRef]
- Li, M.D.; Ruan, H.B.; Hughes, M.E.; Lee, J.S.; Singh, J.P.; Jones, S.P.; Nitabach, M.N.; Yang, X. O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab. 2013, 17, 303–310. [Google Scholar] [CrossRef] [Green Version]
- Berthier, A.; Vinod, M.; Porez, G.; Steenackers, A.; Alexandre, J.; Yamakawa, N.; Gheeraert, C.; Ploton, M.; Marechal, X.; Dubois-Chevalier, J.; et al. Combinatorial regulation of hepatic cytoplasmic signaling and nuclear transcriptional events by the OGT/REV-ERBalpha complex. Proc. Natl. Acad. Sci. USA 2018, 115, E11033–E11042. [Google Scholar] [CrossRef]
- Peek, C.B.; Levine, D.C.; Cedernaes, J.; Taguchi, A.; Kobayashi, Y.; Tsai, S.J.; Bonar, N.A.; McNulty, M.R.; Ramsey, K.M.; Bass, J. Circadian Clock Interaction with HIF1alpha Mediates Oxygenic Metabolism and Anaerobic Glycolysis in Skeletal Muscle. Cell Metab. 2017, 25, 86–92. [Google Scholar] [CrossRef]
- Adamovich, Y.; Ladeuix, B.; Sobel, J.; Manella, G.; Neufeld-Cohen, A.; Assadi, M.H.; Golik, M.; Kuperman, Y.; Tarasiuk, A.; Koeners, M.P.; et al. Oxygen and Carbon Dioxide Rhythms Are Circadian Clock Controlled and Differentially Directed by Behavioral Signals. Cell Metab. 2019. [Google Scholar] [CrossRef]
- Adamovich, Y.; Ladeuix, B.; Golik, M.; Koeners, M.P.; Asher, G. Rhythmic Oxygen Levels Reset Circadian Clocks through HIF1alpha. Cell Metab. 2017, 25, 93–101. [Google Scholar] [CrossRef]
- Rius, J.; Guma, M.; Schachtrup, C.; Akassoglou, K.; Zinkernagel, A.S.; Nizet, V.; Johnson, R.S.; Haddad, G.G.; Karin, M. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature 2008, 453, 807–811. [Google Scholar] [CrossRef] [PubMed]
- Roenneberg, T.; Chua, E.J.; Bernardo, R.; Mendoza, E. Modelling biological rhythms. Curr. Biol. 2008, 18, R826–R835. [Google Scholar] [CrossRef] [PubMed]
- Asgari-Targhi, A.; Klerman, E.B. Mathematical modeling of circadian rhythms. Wiley Interdiscip. Rev. Syst. Biol. Med. 2019, 11, e1439. [Google Scholar] [CrossRef]
- Braun, R.; Kath, W.L.; Iwanaszko, M.; Kula-Eversole, E.; Abbott, S.M.; Reid, K.J.; Zee, P.C.; Allada, R. Universal method for robust detection of circadian state from gene expression. Proc. Natl. Acad. Sci. USA 2018, 115, E9247–E9256. [Google Scholar] [CrossRef]
- Ruben, M.D.; Wu, G.; Smith, D.F.; Schmidt, R.E.; Francey, L.J.; Lee, Y.Y.; Anafi, R.C.; Hogenesch, J.B. A database of tissue-specific rhythmically expressed human genes has potential applications in circadian medicine. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Nohara, K.; Park, N.; Park, Y.S.; Guillory, B.; Zhao, Z.; Garcia, J.M.; Koike, N.; Lee, C.C.; Takahashi, J.S.; et al. The Small Molecule Nobiletin Targets the Molecular Oscillator to Enhance Circadian Rhythms and Protect against Metabolic Syndrome. Cell Metab. 2016, 23, 610–621. [Google Scholar] [CrossRef] [PubMed]


| Protein or Metabolite | Main Peripheral Sources | Main References |
|---|---|---|
| Leptin | Adipose tissue (adipocytes) | [117,125,185,186,187] |
| Kynurenine | Liver (hepatocytes) | [120,188] |
| FGF21 | Liver (hepatocytes) | [189] |
| Ghrelin | Stomach (parietal cells) | [190,191,192,193] |
| NAMPT | Peripheral tissues, adipose tissue | [194,195] |
| Irisin | Skeletal muscle | [120,196,197] |
| Lactate | Skeletal muscle | [198,199] |
| Insulin | Pancreas (β cells) | [185,200,201] |
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maury, E. Off the Clock: From Circadian Disruption to Metabolic Disease. Int. J. Mol. Sci. 2019, 20, 1597. https://doi.org/10.3390/ijms20071597
Maury E. Off the Clock: From Circadian Disruption to Metabolic Disease. International Journal of Molecular Sciences. 2019; 20(7):1597. https://doi.org/10.3390/ijms20071597
Chicago/Turabian StyleMaury, Eleonore. 2019. "Off the Clock: From Circadian Disruption to Metabolic Disease" International Journal of Molecular Sciences 20, no. 7: 1597. https://doi.org/10.3390/ijms20071597
APA StyleMaury, E. (2019). Off the Clock: From Circadian Disruption to Metabolic Disease. International Journal of Molecular Sciences, 20(7), 1597. https://doi.org/10.3390/ijms20071597





