Automated Classification of Colorectal Neoplasms in White-Light Colonoscopy Images via Deep Learning
Abstract
:1. Introduction
2. Materials and Methods
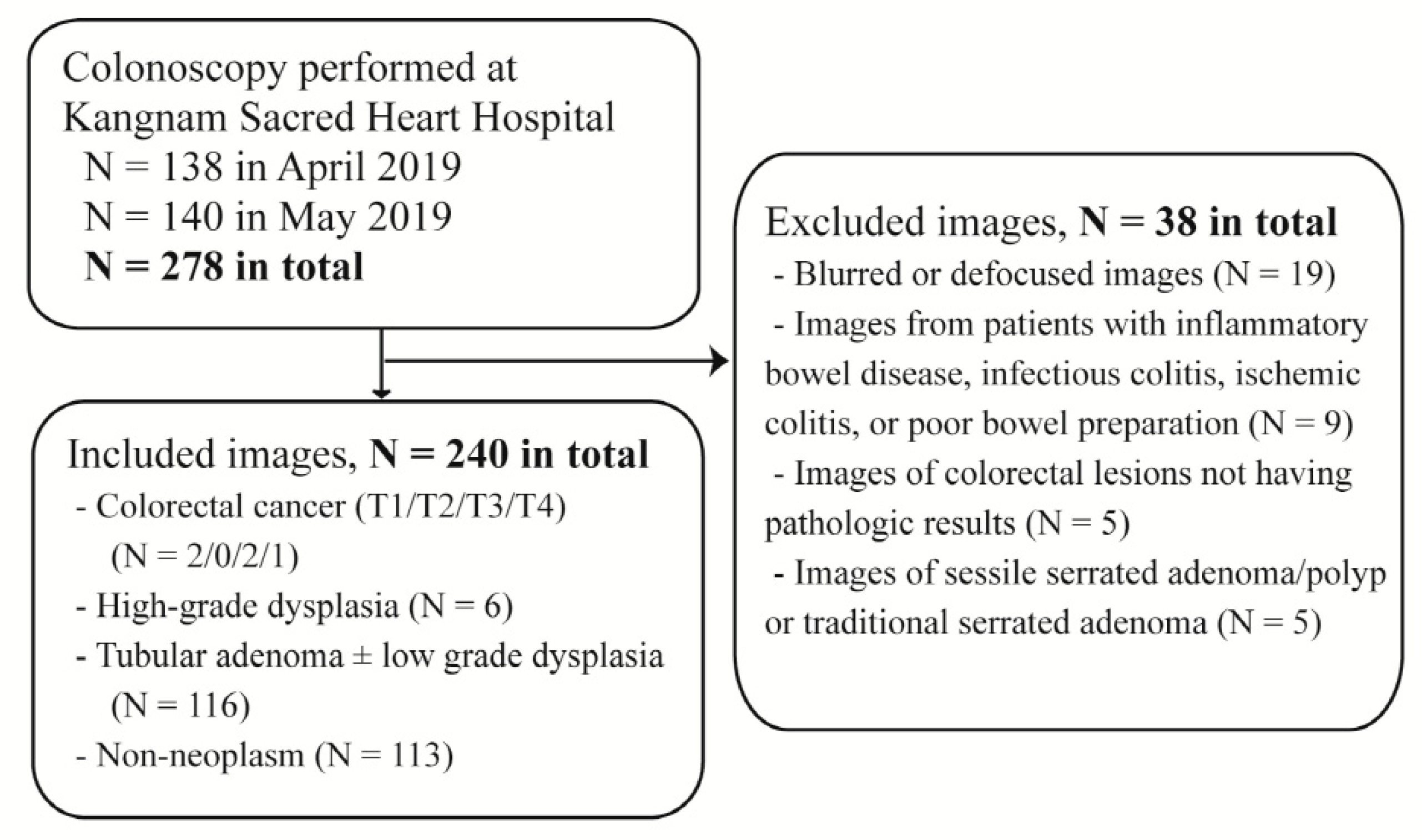
2.1. Data Collection
2.2. Colonoscopy Procedure
2.3. Data Classification
2.4. The Training, Test and External Validation Datasets
2.5. Preprocessing of the Datasets
2.6. Construction of the CNN Models
2.7. Main Outcome Measures
2.8. Statistical Analysis
3. Results
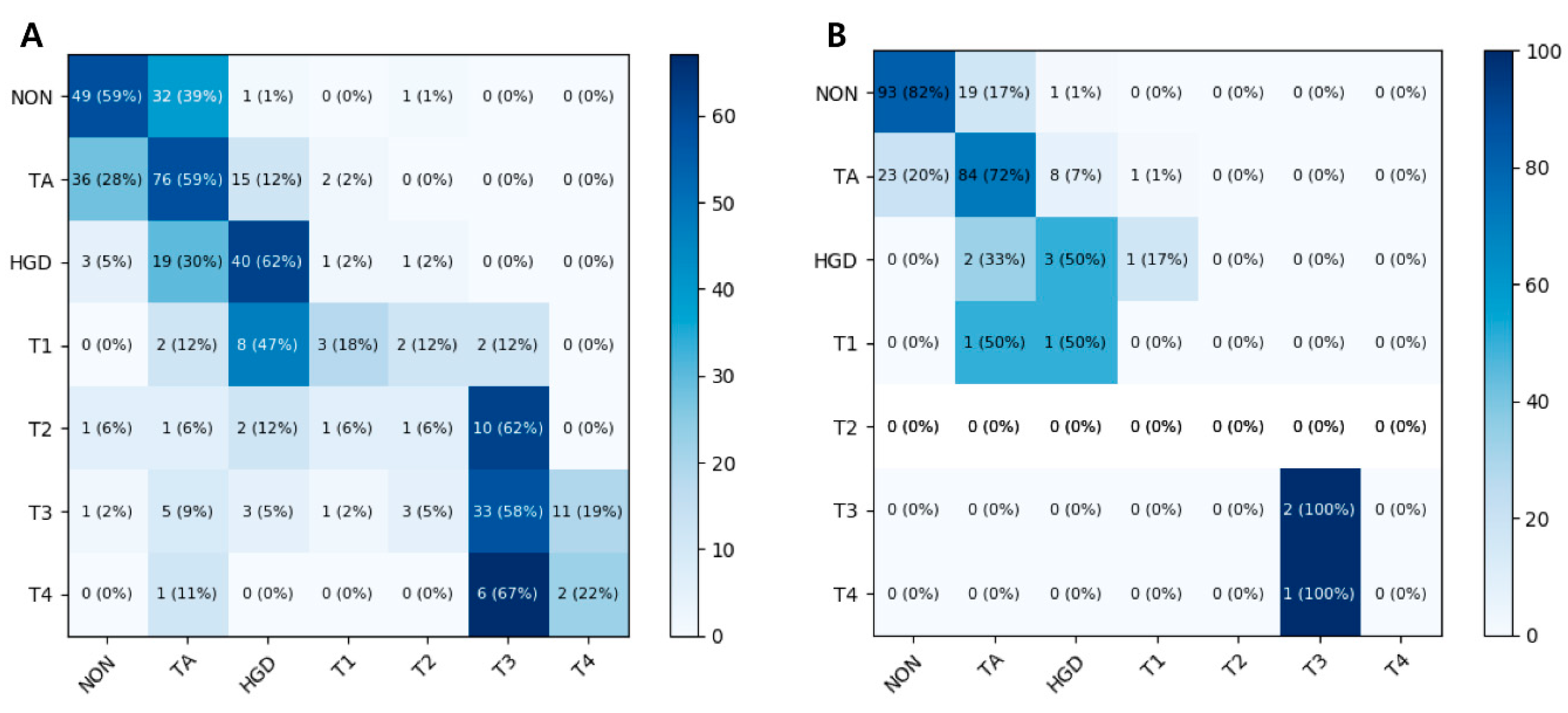
3.1. Seven-Class Classification Performances
3.2. Four-Class Classification Performances
3.3. Binary Classification Performances
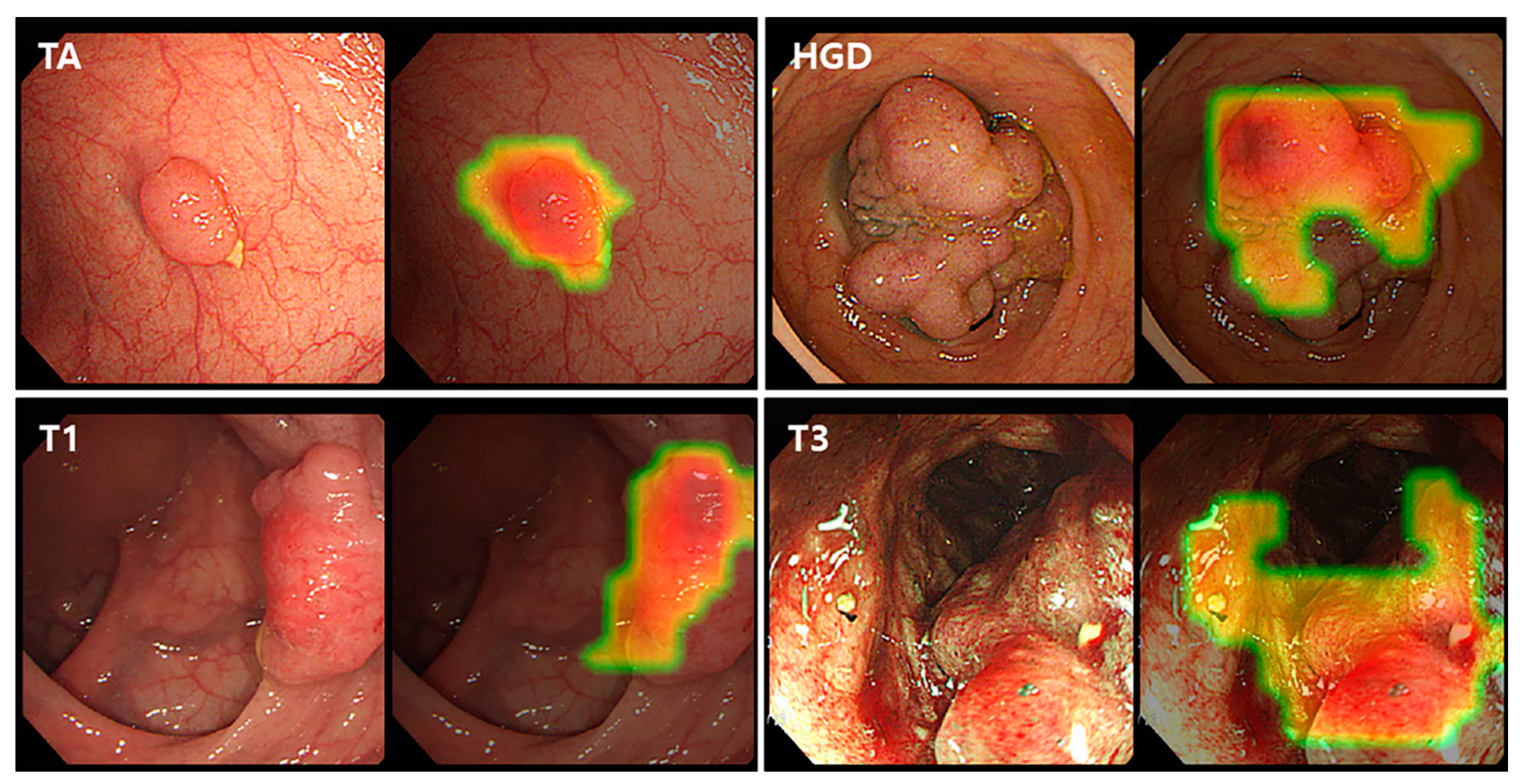
3.4. Class Activation Map
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, K.-W.; Won, Y.-J.; Kong, H.-J.; Oh, C.-M.; Cho, H.; Lee, D.H.; Lee, K.H. Cancer Statistics in Korea: Incidence, Mortality, Survival, and Prevalence in 2012. Cancer Res. Treat 2015, 47, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Zauber, A.G.; Winawer, S.J.; O’Brien, M.J.; Lansdorp-Vogelaar, I.; Van Ballegooijen, M.; Hankey, B.F.; Shi, W.; Bond, J.H.; Schapiro, M.; Panish, J.F.; et al. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N. Engl. J. Med. 2012, 366, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Rex, D.K.; Boland, R.C.; Dominitz, J.A.; Giardiello, F.M.; A Johnson, D.; Kaltenbach, T.; Levin, T.R.; Lieberman, D.; Robertson, D.J.; Boland, C.R. Colorectal Cancer Screening: Recommendations for Physicians and Patients from the U.S. Multi-Society Task Force on Colorectal Cancer. Am. J. Gastroenterol. 2017, 112, 1016–1030. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.I.; Hong, S.P.; Kim, S.E.; Kim, H.S.; Yang, D.H.; Shin, S.J.; Lee, S.H.; Kim, Y.H.; Park, D.I.; Yang, S.K.; et al. Korean Guidelines for Colorectal Cancer Screening and Polyp Detection. J. Korean Soc. Radiol. 2012, 66, 385. [Google Scholar] [CrossRef] [Green Version]
- Hassan, C.; Pickhardt, P.J.; Rex, U.K. A Resect and Discard Strategy Would Improve Cost-Effectiveness of Colorectal Cancer Screening. Clin. Gastroenterol. Hepatol. 2010, 8, 865–869.e3. [Google Scholar] [CrossRef]
- Peery, A.F.; Cools, K.S.; Strassle, P.D.; McGill, S.K.; Crockett, S.D.; Barker, A.; Koruda, M.; Grimm, I.S. Increasing Rates of Surgery for Patients With Nonmalignant Colorectal Polyps in the United States. Gastroenterology 2018, 154, 1352–1360.e3. [Google Scholar] [CrossRef]
- Keswani, R.N.; Law, R.; Ciolino, J.D.; Lo, A.A.; Gluskin, A.B.; Bentrem, D.J.; Komanduri, S.; Pacheco, J.; Grande, D.; Thompson, W.K. Adverse events after surgery for nonmalignant colon polyps are common and associated with increased length of stay and costs. Gastrointest Endosc. 2016, 84, 296–303.e1. [Google Scholar] [CrossRef]
- East, J.; Vleugels, J.L.; Roelandt, P.; Bhandari, P.; Bisschops, R.; Dekker, E.; Hassan, C.; Horgan, G.; Kiesslich, R.; Longcroft-Wheaton, G.; et al. Advanced endoscopic imaging: European Society of Gastrointestinal Endoscopy (ESGE) Technology Review. Endoscopy 2016, 48, 1029–1045. [Google Scholar] [CrossRef] [Green Version]
- Hewett, D.G.; Kaltenbach, T.; Sano, Y.; Tanaka, S.; Saunders, B.P.; Ponchon, T.; Soetikno, R.; Rex, D.K. Validation of a Simple Classification System for Endoscopic Diagnosis of Small Colorectal Polyps Using Narrow-Band Imaging. Gastroenterology 2012, 143, 599–607.e1. [Google Scholar] [CrossRef]
- Wanders, L.K.; E East, J.; E Uitentuis, S.; Leeflang, M.; Dekker, E. Diagnostic performance of narrowed spectrum endoscopy, autofluorescence imaging, and confocal laser endomicroscopy for optical diagnosis of colonic polyps: A meta-analysis. Lancet Oncol. 2013, 14, 1337–1347. [Google Scholar] [CrossRef]
- Kuiper, T.; Marsman, W.A.; Jansen, J.M.; Van Soest, E.J.; Haan, Y.C.; Bakker, G.J.; Fockens, P.; Dekker, E. Accuracy for Optical Diagnosis of Small Colorectal Polyps in Nonacademic Settings. Clin. Gastroenterol. Hepatol. 2012, 10, 1016–1020. [Google Scholar] [CrossRef] [PubMed]
- Ladabaum, U.; Fioritto, A.; Mitani, A.; Desai, M.; Kim, J.P.; Rex, U.K.; Imperiale, T.; Gunaratnam, N. Real-time optical biopsy of colon polyps with narrow band imaging in community practice does not yet meet key thresholds for clinical decisions. Gastroenterology 2012, 144, 81–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 2016, 316, 2402–2410. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.J.; Bang, C.S. Application of artificial intelligence in gastroenterology. World J. Gastroenterol. 2019, 25, 1666–1683. [Google Scholar] [CrossRef] [PubMed]
- Cho, B.-J.; Bang, C.S.; Park, S.W.; Yang, Y.J.; Seo, S.I.; Lim, H.; Shin, W.G.; Hong, J.T.; Yoo, Y.T.; Hong, S.H.; et al. Automated classification of gastric neoplasms in endoscopic images using a convolutional neural network. Endoscopy 2019, 51, 1121–1129. [Google Scholar] [CrossRef]
- Urban, G.; Tripathi, P.; Alkayali, T.; Mittal, M.; Jalali, F.; Karnes, W.; Baldi, P. Deep Learning Localizes and Identifies Polyps in Real Time With 96% Accuracy in Screening Colonoscopy. Gastroenterology 2018, 155, 1069–1078.e8. [Google Scholar] [CrossRef]
- Klare, P.; Sander, C.; Prinzen, M.; Haller, B.; Nowack, S.; Abdelhafez, M.; Poszler, A.; Brown, H.; Wilhelm, D.; Schmid, R.M.; et al. Automated polyp detection in the colorectum: A prospective study (with videos). Gastrointest Endosc. 2019, 89, 576–582.e1. [Google Scholar] [CrossRef]
- Wang, P.; Berzin, T.M.; Brown, J.R.G.; Bharadwaj, S.; Becq, A.; Xiao, X.; Liu, P.; Li, L.; Song, Y.; Zhang, D.; et al. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: A prospective randomised controlled study. Gut 2019, 68, 1813–1819. [Google Scholar] [CrossRef] [Green Version]
- Byrne, M.F.; Chapados, N.; Soudan, F.; Oertel, C.; Pérez, M.L.; Kelly, R.; Iqbal, N.; Chandelier, F.; Rex, D.K. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut 2017, 68, 94–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, P.-J.; Lin, M.-C.; Lai, M.-J.; Lin, J.-C.; Lu, H.H.-S.; Tseng, V.S. Accurate Classification of Diminutive Colorectal Polyps Using Computer-Aided Analysis. Gastroenterology 2018, 154, 568–575. [Google Scholar] [CrossRef] [PubMed]
- Komeda, Y.; Handa, H.; Watanabe, T.; Nomura, T.; Kitahashi, M.; Sakurai, T.; Okamoto, A.; Minami, T.; Kono, M.; Arizumi, T.; et al. Computer-Aided Diagnosis Based on Convolutional Neural Network System for Colorectal Polyp Classification: Preliminary Experience. Oncology 2017, 93, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Mori, Y.; Kudo, S.-E.; Misawa, M.; Saito, Y.; Ikematsu, H.; Hotta, K.; Ohtsuka, K.; Urushibara, F.; Kataoka, S.; Ogawa, Y.; et al. Real-Time Use of Artificial Intelligence in Identification of Diminutive Polyps During Colonoscopy. Ann. Intern. Med. 2018, 169, 357. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity Mappings in Deep Residual Networks. arXiv 2016, arXiv:1603.05027. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. arXiv 2016, arXiv:1602.07261. [Google Scholar]
- Huang, G.; Li, Y.; Pleiss, G.; Liu, Z.; Hopcroft, J.E.; Weinberger, K.Q. Snapshot Ensembles: Train 1, get M for free. arXiv 2017, arXiv:1704.00109. [Google Scholar]
- Hayashi, N.; Tanaka, S.; Hewett, D.G.; Kaltenbach, T.R.; Sano, Y.; Ponchon, T.; Saunders, B.P.; Rex, U.K.; Soetikno, R.M. Endoscopic prediction of deep submucosal invasive carcinoma: Validation of the Narrow-Band Imaging International Colorectal Endoscopic (NICE) classification. Gastrointest. Endosc. 2013, 78, 625–632. [Google Scholar] [CrossRef]
- Sano, Y.; Tanaka, S.; Kudo, S.-E.; Saito, S.; Matsuda, T.; Wada, Y.; Fujii, T.; Ikematsu, H.; Uraoka, T.; Kobayashi, N.; et al. Narrow-band imaging (NBI) magnifying endoscopic classification of colorectal tumors proposed by the Japan NBI Expert Team. Dig. Endosc. 2016, 28, 526–533. [Google Scholar] [CrossRef]
- Zhang, R.; Mak, T.W.C.; Wong, S.H.; Lau, J.Y.W.; Poon, C.C.Y.; Zheng, Y.; Yu, R. Automatic Detection and Classification of Colorectal Polyps by Transferring Low-Level CNN Features From Nonmedical Domain. IEEE J. Biomed. Health Inform. 2016, 21, 41–47. [Google Scholar] [CrossRef]
- Chao, W.-L.; Manickavasagan, H.; Krishna, S.G. Application of Artificial Intelligence in the Detection and Differentiation of Colon Polyps: A Technical Review for Physicians. Diagnostics 2019, 9, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, Y.; Kudo, S.-E.; Berzin, T.M.; Misawa, M.; Takeda, K. Computer-aided diagnosis for colonoscopy. Endoscopy 2017, 49, 813–819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, K.; Kudo, S.-E.; Mori, Y.; Misawa, M.; Kudo, T.; Wakamura, K.; Katagiri, A.; Baba, T.; Hidaka, E.; Ishida, F.; et al. Accuracy of diagnosing invasive colorectal cancer using computer-aided endocytoscopy. Endoscopy 2017, 49, 798–802. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Montes, C.; Sánchez, F.J.; Bernal, J.; Córdova, H.; Lopez-Ceron, M.; Cuatrecasas, M.; De Miguel, C.R.; García-Rodríguez, A.; Durán, R.G.; Pellisé, M.; et al. Computer-aided prediction of polyp histology on white light colonoscopy using surface pattern analysis. Endoscopy 2018, 51, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Rex, D.K.; Kahi, C.; O’Brien, M.J.; Levin, T.; Pohl, H.; Rastogi, A.; Burgart, L.; Imperiale, T.; Ladabaum, U.; Cohen, J.; et al. The American Society for Gastrointestinal Endoscopy PIVI (Preservation and Incorporation of Valuable Endoscopic Innovations) on real-time endoscopic assessment of the histology of diminutive colorectal polyps. Gastrointest Endosc. 2011, 73, 419–422. [Google Scholar] [CrossRef]
- Park, H.-W.; Byeon, J.; Yang, S.-K.; Kim, H.S.; Kim, W.; Kim, T.I.; Park, N.I.; Kim, Y.-H.; Kim, H.J.; Lee, M.S.; et al. Colorectal Neoplasm in Asymptomatic Average-risk Koreans: The KASID Prospective Multicenter Colonoscopy Survey. Gut Liver 2009, 3, 35–40. [Google Scholar] [CrossRef] [Green Version]


| Model | Diagnostic Performance,% (95% CI) | AUC (95% CI) | ||||
|---|---|---|---|---|---|---|
| Accuracy (%) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | ||
| Neoplastic lesions vs. non-neoplastic lesions | ||||||
| ResNet-152 | 79.4 (78.5–80.3) | 95.4 (93.2–97.6) | 30.1 (25.5–34.7) | 80.8 (78.4–83.2) | 68.8 (58.4–79.2) | 0.821 (0.802–0.840) |
| Inception-ResNet-v2 | 79.5 (77.6–81.4) | 94.1 (92.5–95.7) | 34.1 (28.1–40.1) | 81.6 (80.6–82.6) | 65.0 (54.7–75.3) | 0.832 (0.810–0.854) |
| Advanced colorectal lesions vs. non-advanced colorectal lesions | ||||||
| ResNet-152 | 86.7 (84.9–88.5) | 80.0 (75.4–84.6) | 91.3 (90.8–91.8) | 86.0 (83.7–88.3) | 87.1 (85.1–89.1) | 0.929 (0.927–0.931) |
| Inception-ResNet-v2 | 87.1 (86.2–88.0) | 83.2 (81.5–84.9) | 89.7 (87.7–91.7) | 84.5 (81.0–88.0) | 88.7 (87.7–89.7) | 0.935 (0.929–0.941) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.J.; Cho, B.-J.; Lee, M.-J.; Kim, J.H.; Lim, H.; Bang, C.S.; Jeong, H.M.; Hong, J.T.; Baik, G.H. Automated Classification of Colorectal Neoplasms in White-Light Colonoscopy Images via Deep Learning. J. Clin. Med. 2020, 9, 1593. https://doi.org/10.3390/jcm9051593
Yang YJ, Cho B-J, Lee M-J, Kim JH, Lim H, Bang CS, Jeong HM, Hong JT, Baik GH. Automated Classification of Colorectal Neoplasms in White-Light Colonoscopy Images via Deep Learning. Journal of Clinical Medicine. 2020; 9(5):1593. https://doi.org/10.3390/jcm9051593
Chicago/Turabian StyleYang, Young Joo, Bum-Joo Cho, Myung-Je Lee, Ju Han Kim, Hyun Lim, Chang Seok Bang, Hae Min Jeong, Ji Taek Hong, and Gwang Ho Baik. 2020. "Automated Classification of Colorectal Neoplasms in White-Light Colonoscopy Images via Deep Learning" Journal of Clinical Medicine 9, no. 5: 1593. https://doi.org/10.3390/jcm9051593
APA StyleYang, Y. J., Cho, B.-J., Lee, M.-J., Kim, J. H., Lim, H., Bang, C. S., Jeong, H. M., Hong, J. T., & Baik, G. H. (2020). Automated Classification of Colorectal Neoplasms in White-Light Colonoscopy Images via Deep Learning. Journal of Clinical Medicine, 9(5), 1593. https://doi.org/10.3390/jcm9051593






