Molecular Diversity and Combining Ability in Newly Developed Maize Inbred Lines under Low-Nitrogen Conditions
Abstract
:1. Introduction
2. Materials and Methods
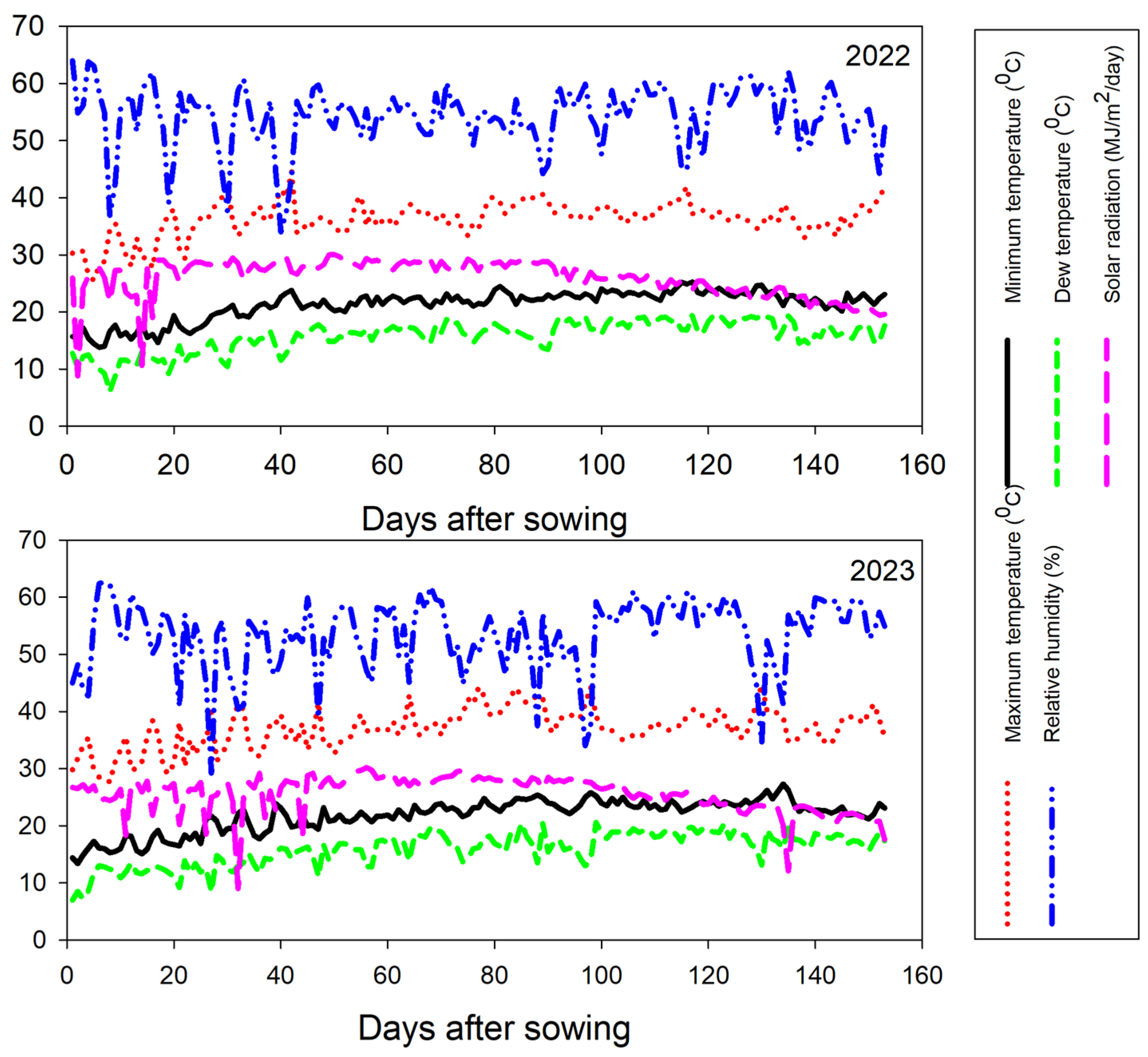
2.1. Plant Materials and Experimental Design
2.2. Data Collection
2.3. Statistical Analysis
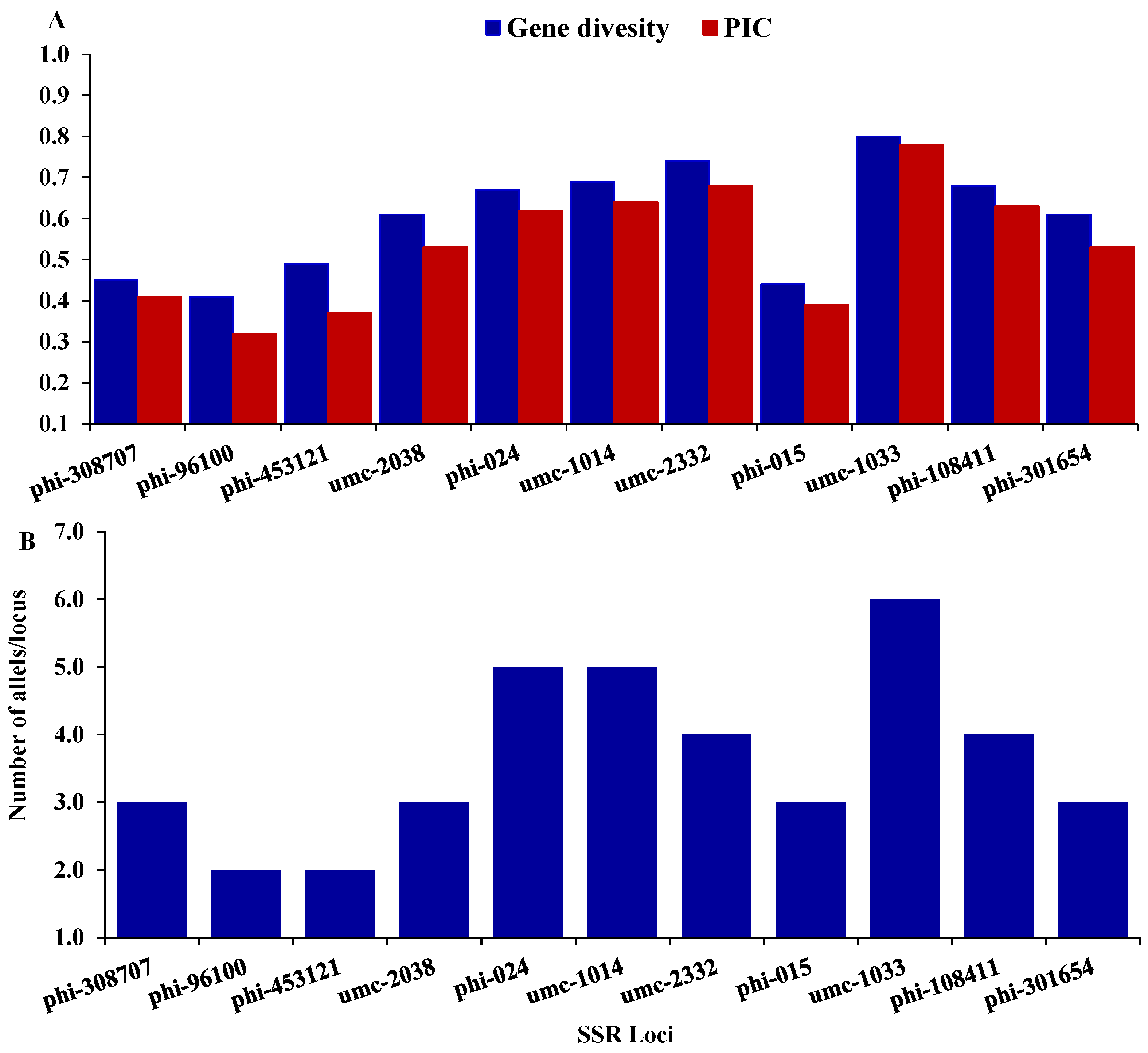
2.4. Molecular Analysis
3. Results
3.1. Genetic Diversity among Inbred Lines
3.2. Analysis of Variance
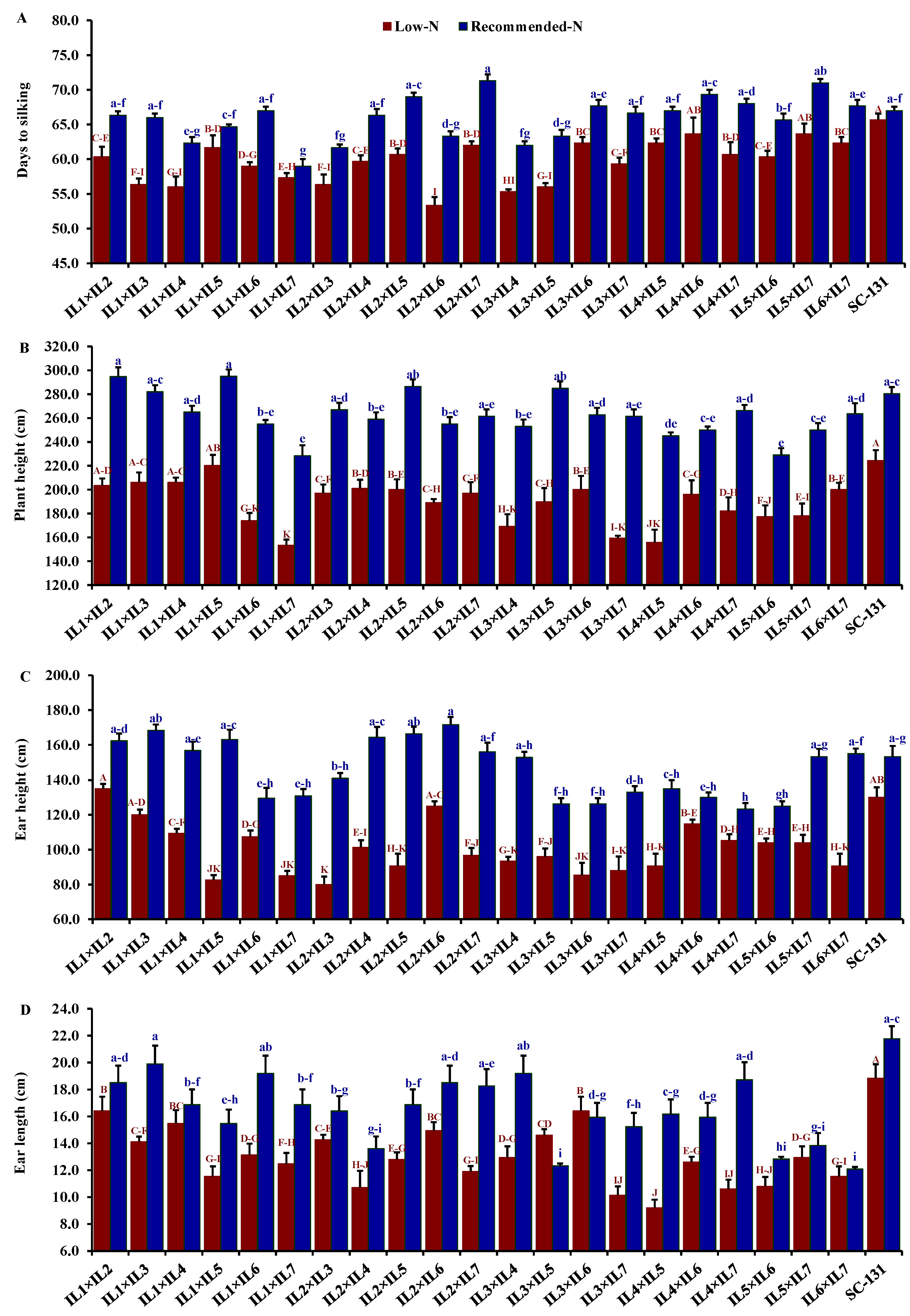
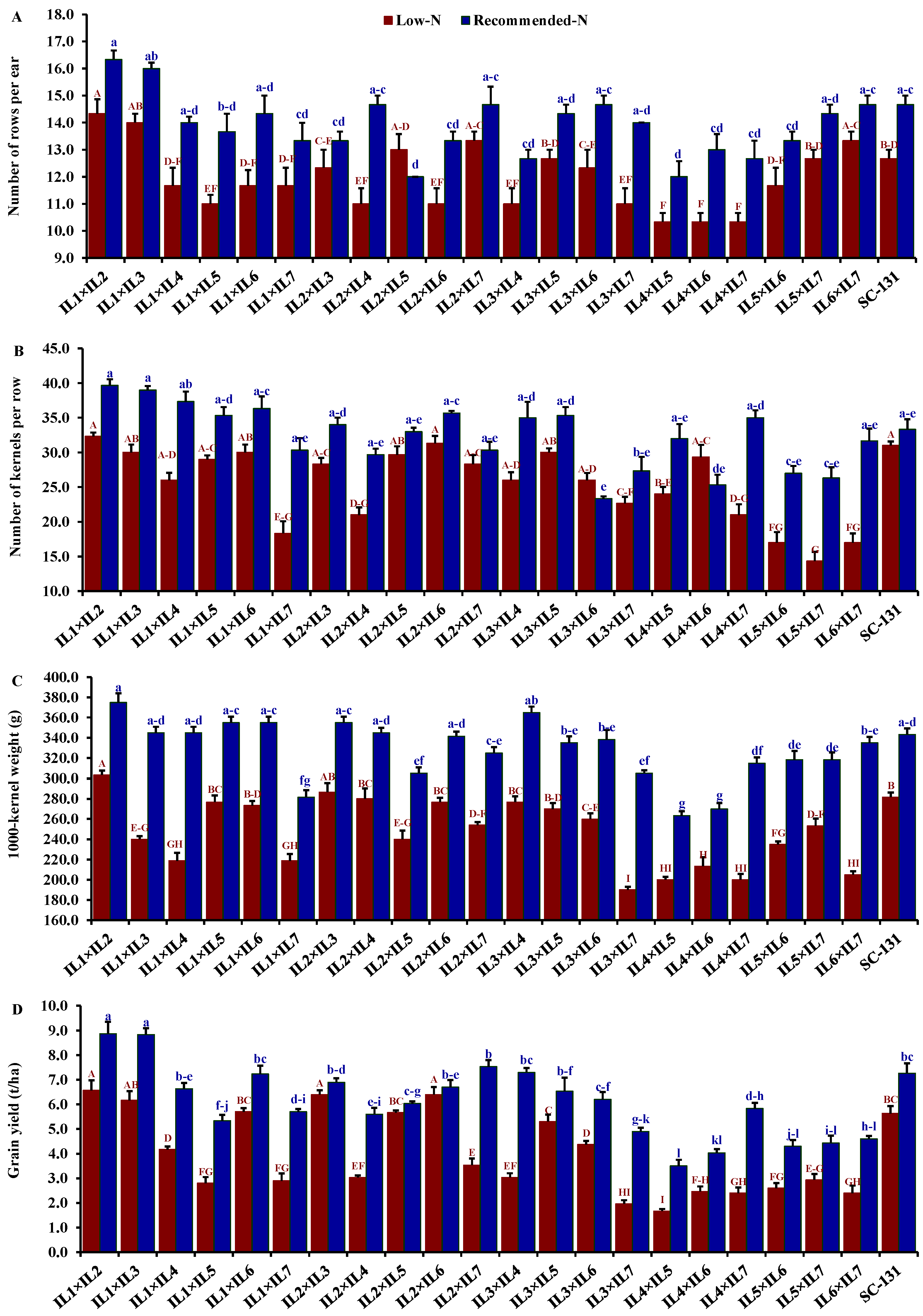
3.3. Performance of the Evaluated Hybrids
3.4. Classification of Hybrids
3.5. General Combining Ability (GCA)
3.6. Specific Combining Ability (SCA)
3.7. Interrelationship among Evaluated Hybrids and Measured Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. Food and Agriculture Organization of the United Nations. Statistical Database. 2024. Available online: http://www.fao.org/faostat/en/#data (accessed on 3 January 2024).
- Wang, H.; Ren, H.; Zhang, L.; Zhao, Y.; Liu, Y.; He, Q.; Li, G.; Han, K.; Zhang, J.; Zhao, B. A sustainable approach to narrowing the summer maize yield gap experienced by smallholders in the North China Plain. Agric. Syst. 2023, 204, 103541. [Google Scholar] [CrossRef]
- Mathiang, E.A.; Sa, K.J.; Park, H.; Kim, Y.J.; Lee, J.K. Genetic Diversity and Population Structure of Normal Maize Germplasm Collected in South Sudan Revealed by SSR Markers. Plants 2022, 11, 2787. [Google Scholar] [CrossRef]
- Mukri, G.; Patil, M.S.; Motagi, B.N.; Bhat, J.S.; Singh, C.; Jeevan Kumar, S.; Gadag, R.; Gupta, N.C.; Simal-Gandara, J. Genetic variability, combining ability and molecular diversity-based parental line selection for heterosis breeding in field corn (Zea mays L.). Mol. Biol. Rep. 2022, 49, 4517–4524. [Google Scholar] [CrossRef]
- Bakala, H.S.; Devi, J.; Ankita; Sarao, L.K.; Kaur, S. Utilization of wheat and maize waste as biofuel source. In Agroindustrial Waste for Green Fuel Application; Clean Energy Production Technologies; Springer: Singapore, 2023; pp. 27–66. [Google Scholar] [CrossRef]
- Banaj, A.; Banaj, Đ.; Stipešević, B.; Nemet, F. Seeding pattern impact at crop density establishment and grain yield of maize. Crops 2023, 3, 1–10. [Google Scholar] [CrossRef]
- Liu, J.; You, L.; Amini, M.; Obersteiner, M.; Herrero, M.; Zehnder, A.J.; Yang, H. A high-resolution assessment on global nitrogen flows in cropland. Proc. Natl. Acad. Sci. USA 2010, 107, 8035–8040. [Google Scholar] [CrossRef]
- Santos, T.d.O.; Amaral Junior, A.T.d.; Moulin, M.M. Maize breeding for low nitrogen inputs in agriculture: Mechanisms underlying the tolerance to the abiotic stress. Stresses 2023, 3, 136–152. [Google Scholar] [CrossRef]
- Fageria, N.K.; Baligar, V. Enhancing nitrogen use efficiency in crop plants. Adv. Agron. 2005, 88, 97–185. [Google Scholar] [CrossRef]
- Mu, X.; Chen, Y. The physiological response of photosynthesis to nitrogen deficiency. Plant Physiol. Biochem. 2021, 158, 76–82. [Google Scholar] [CrossRef]
- Leghari, S.J.; Wahocho, N.A.; Laghari, G.M.; HafeezLaghari, A.; MustafaBhabhan, G.; HussainTalpur, K.; Bhutto, T.A.; Wahocho, S.A.; Lashari, A.A. Role of nitrogen for plant growth and development: A review. Adv. Environ. Biol. 2016, 10, 209–219. [Google Scholar]
- Ciampitti, I.A.; Vyn, T. Physiological perspectives of changes over time in maize yield dependency on nitrogen uptake and associated nitrogen efficiencies: A review. Field Crop. Res. 2012, 133, 48–67. [Google Scholar] [CrossRef]
- Mayer, L.I.; Rossini, M.d.L.A.; Maddonni, G.A. Inter-plant variation of grain yield components and kernel composition of maize crops grown under contrasting nitrogen supply. Field Crop. Res. 2012, 125, 98–108. [Google Scholar] [CrossRef]
- Okunlola, G.; Badu-Apraku, B.; Ariyo, O.; Ayo-Vaughan, M. The combining ability of extra-early maturing quality protein maize (Zea mays) inbred lines and the performance of their hybrids in Striga-infested and low-nitrogen environments. Front. Sustain. Food Syst. 2023, 7, 1238874. [Google Scholar] [CrossRef]
- Annor, B.; Badu-Apraku, B. Selection of extra-early white quality protein maize (Zea mays L.) inbred lines for drought and low soil nitrogen resilient hybrid production. Front. Sustain. Food Syst. 2024, 7, 1238776. [Google Scholar] [CrossRef]
- Chen, Y.; Xiao, C.; Wu, D.; Xia, T.; Chen, Q.; Chen, F.; Yuan, L.; Mi, G. Effects of nitrogen application rate on grain yield and grain nitrogen concentration in two maize hybrids with contrasting nitrogen remobilization efficiency. Eur. J. Agron. 2015, 62, 79–89. [Google Scholar] [CrossRef]
- Costa, C.; Dwyer, L.M.; Stewart, D.W.; Smith, D.L. Nitrogen effects on grain yield and yield components of leafy and nonleafy maize genotypes. Crop Sci. 2002, 42, 1556–1563. [Google Scholar] [CrossRef]
- Hou, P.; Gao, Q.; Xie, R.; Li, S.; Meng, Q.; Kirkby, E.A.; Römheld, V.; Müller, T.; Zhang, F.; Cui, Z. Optimizing nitrogen management for spring maize grown in China. Field Crop. Res. 2012, 129, 1–6. [Google Scholar] [CrossRef]
- Gharib, M.A.A.H.; Qabil, N.; Salem, A.H.; Ali, M.M.A.; Awaad, H.A.; Mansour, E. Characterization of wheat landraces and commercial cultivars based on morpho-phenological and agronomic traits. Cereal Res. Commun. 2020, 49, 149–159. [Google Scholar] [CrossRef]
- Zannat, A.; Hussain, M.A.; Abdullah, A.H.M.; Hossain, M.I.; Saifullah, M.; Safhi, F.A.; Alshallash, K.S.; Mansour, E.; ElSayed, A.I.; Hossain, M.S. Exploring genotypic variability and interrelationships among growth, yield, and quality characteristics in diverse tomato genotypes. Heliyon 2023, 9, e18958. [Google Scholar] [CrossRef]
- Gracia, M.P.; Mansour, E.; Casas, A.M.; Lasa, J.M.; Medina, B.; Molina-Cano, J.L.; Moralejo, M.A.; López, A.; López-Fuster, P.; Escribano, J.; et al. Progress in the Spanish national barley breeding program. Span. J. Agric. Res. 2012, 10, 741. [Google Scholar] [CrossRef]
- Siddu, C.B.; Ramesh, S.; Kalpana, M.P.; Basanagouda, G.; Sathish, H.; Aniketh, S.; Karthik, N.; Sindhu, D.; Kemparaju, M.; Kirankumar, R. Identification of inbred lines harbouring favourable dominant alleles not present in the parents of three elite maize (Zea mays L.) single cross hybrids. Genet. Resour. Crop Evol. 2024, 71, 851–862. [Google Scholar] [CrossRef]
- Riyanto, A.; Hidayat, P.; Suprayogi, Y.; Haryanto, T.A.D. Diallel analysis of length and shape of rice using Hayman and Griffing method. Open Agric. 2023, 8, 20220169. [Google Scholar] [CrossRef]
- Kamara, M.M.; Rehan, M.; Mohamed, A.M.; El Mantawy, R.F.; Kheir, A.M.S.; Abd El-Moneim, D.; Safhi, F.A.; Alshamrani, S.M.; Hafez, E.M.; Behiry, S.I.; et al. Genetic potential and inheritance patterns of physiological, agronomic and quality traits in bread wheat under normal and water deficit conditions. Plants 2022, 11, 952. [Google Scholar] [CrossRef]
- Fan, X.; Zhang, Y.; Yao, W.; Bi, Y.; Liu, L.; Chen, H.; Kang, M. Reciprocal diallel crosses impact combining ability, variance estimation, and heterotic group classification. Crop Sci. 2014, 54, 89–97. [Google Scholar] [CrossRef]
- Salem, T.; Rabie, H.; Mowafy, S.; Eissa, A.; Mansour, E. Combining ability and genetic components of egyptian cotton for earliness, yield, and fiber quality traits. SABRAO J. Breed. Genet. 2020, 52, 369–389. [Google Scholar]
- Heiba, H.; Mahgoub, E.; Mahmoud, A.; Ibrahİm, M.; Mansour, E. Combining ability and gene action controlling chocolate spot resistance and yield traits in faba bean (Vicia faba L.). J. Agric. Sci. 2023, 29, 77–88. [Google Scholar] [CrossRef]
- ElShamey, E.A.Z.; Sakran, R.M.; ElSayed, M.A.A.; Aloufi, S.; Alharthi, B.; Alqurashi, M.; Mansour, E.; Abd El-Moneim, D. Heterosis and combining ability for floral and yield characters in rice using cytoplasmic male sterility system. Saudi J. Biol. Sci. 2022, 29, 3727–3738. [Google Scholar] [CrossRef]
- Sakran, R.M.; Ghazy, M.I.; Rehan, M.; Alsohim, A.S.; Mansour, E. Molecular genetic diversity and combining ability for some physiological and agronomic traits in rice under well-watered and water-deficit conditions. Plants 2022, 11, 702. [Google Scholar] [CrossRef]
- Hallauer, A.R.; Carena, M.J.; Miranda Filho, J.B. Quantitative Genetics in Maize Breeding, 3rd ed.; Springer: Dordrecht/Heidelberg, Germany; London, UK, 2010; p. 663. [Google Scholar]
- Al-Naggar, A.; El-Ganayni, A.; El-Lakany, M.; El-Sherbeiny, H.; Soliman, M. Mode of inheritance of maize resistance to the pink stem borer, Sesamia cretica Led. under artificial infestation. Egypt. J. Plant Breed. 2000, 4, 13–35. [Google Scholar] [CrossRef]
- Owusu, G.; Nyadanu, D.; Obeng-Antwi, K.; Amoah, R.A.; Danso, F.; Amissah, S. Estimating gene action, combining ability and heterosis for grain yield and agronomic traits in extra-early maturing yellow maize single-crosses under three agro-ecologies of Ghana. Euphytica 2017, 213, 287. [Google Scholar] [CrossRef]
- Shirinpour, M.; Atazadeh, E.; Bybordi, A.; Monirifar, H.; Amini, A.; Hossain, M.A.; Aharizad, S.; Asghari, A. Gene action and inheritance of grain yield and root morphological traits in hybrid maize grown under water deficit conditions. S. Afr. J. Bot. 2023, 161, 180–191. [Google Scholar] [CrossRef]
- Meseka, S.; Menkir, A.; Ibrahim, A.; Ajala, S. Genetic analysis of performance of maize inbred lines selected for tolerance to drought under low nitrogen. Maydica 2006, 51, 487–495. [Google Scholar]
- Noëlle, M.A.H.; Richard, K.; Vernon, G.; Martin, Y.A.; Laouali, M.N.; Liliane, T.N.; Godswill, N.-N. Combining ability and gene action of tropical maize (Zea mays L.) inbred lines under low and high nitrogen conditions. J. Agric. Sci. 2017, 9, 222–235. [Google Scholar] [CrossRef]
- Makumbi, D.; Betrán, J.F.; Bänziger, M.; Ribaut, J.-M. Combining ability, heterosis and genetic diversity in tropical maize (Zea mays L.) under stress and non-stress conditions. Euphytica 2011, 180, 143–162. [Google Scholar] [CrossRef]
- Badu-Apraku, B.; Fakorede, M.; Talabi, A.; Oyekunle, M.; Akaogu, I.; Akinwale, R.; Annor, B.; Melaku, G.; Fasanmade, Y.; Aderounmu, M. Gene action and heterotic groups of early white quality protein maize inbreds under multiple stress environments. Crop Sci. 2016, 56, 183–199. [Google Scholar] [CrossRef]
- Ribeiro, P.; Apraku, B.B.; Gracen, V.; Danquah, E.; Afriyie-Debrah, C.; Obeng-Dankwa, K.; Toyinbo, J. Combining ability and testcross performance of low N tolerant intermediate maize inbred lines under low soil nitrogen and optimal environments. J. Agric. Sci. 2020, 158, 351–370. [Google Scholar] [CrossRef]
- Kasoma, C.; Shimelis, H.; Laing, M.D.; Shayanowako, A.I.; Mathew, I. Revealing the genetic diversity of maize (Zea mays L.) populations by phenotypic traits and DArTseq markers for variable resistance to fall armyworm. Genet. Resour. Crop Evol. 2021, 68, 243–259. [Google Scholar] [CrossRef]
- Iqbal, J.; Shinwari, Z.K.; Rabbani, M.A.; Khan, S.A. Genetic divergence in maize (Zea mays L.) germplasm using quantitative and qualitative traits. Pak. J. Bot. 2015, 47, 227–238. [Google Scholar]
- Schad, P. World Reference Base for Soil Resources—Its fourth edition and its history. J. Plant Nutr. Soil Sci. 2023, 186, 151–163. [Google Scholar] [CrossRef]
- Griffing, B. Concept of general and specific combining ability in relation to diallel crossing systems. Aust. J. Biol. Sci. 1956, 9, 463–493. [Google Scholar] [CrossRef]
- Doyle, J. A rapid total DNA preparation procedure for fresh plant tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Jaccard, P. Nouvelles recherches sur la distribution florale. Bull. Soc. Vaud. Sci. Nat. 1908, 44, 223–270. [Google Scholar] [CrossRef]
- Riache, M.; Revilla, P.; Maafi, O.; Malvar, R.A.; Djemel, A. Combining ability and heterosis of algerian saharan maize populations (Zea mays L.) for tolerance to no-nitrogen fertilization and drought. Agronomy 2021, 11, 492. [Google Scholar] [CrossRef]
- Makore, F.; Magorokosho, C.; Dari, S.; Gasura, E.; Mazarura, U.; Kamutando, C.N. Genetic potential of new maize inbred lines in single-cross hybrid combinations under low-nitrogen stress and optimal conditions. Agronomy 2022, 12, 2205. [Google Scholar] [CrossRef]
- Mebratu, A.; Wegary, D.; Mohammed, W.; Teklewold, A.; Tarekegne, A. Genotype× environment interaction of quality protein maize hybrids under contrasting management conditions in Eastern and Southern Africa. Crop Sci. 2019, 59, 1576–1589. [Google Scholar] [CrossRef]
- Dosho, B.M.; Ifie, B.; Asante, I.; Danquah, E.; Zeleke, H. Genotype-by-environment interaction and yield stability for grain yield of quality protein maize hybrids under low and optimum soil nitrogen environments. J. Crop Sci. Biotech. 2022, 25, 437–450. [Google Scholar] [CrossRef]
- Xu, G.; Fan, X.; Miller, A.J.A.r.o.p.b. Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 2012, 63, 153–182. [Google Scholar] [CrossRef]
- Uribelarrea, M.; Crafts-Brandner, S.J.; Below, F.E. Physiological N response of field-grown maize hybrids (Zea mays L.) with divergent yield potential and grain protein concentration. Plant Soil 2009, 316, 151–160. [Google Scholar] [CrossRef]
- Kant, S.; Bi, Y.-M.; Rothstein, S. Understanding plant response to nitrogen limitation for the improvement of crop nitrogen use efficiency. J. Exp. Bot. 2011, 62, 1499–1509. [Google Scholar] [CrossRef]
- Wu, Y.-w.; Qiang, L.; Rong, J.; Wei, C.; Liu, X.-L.; Kong, F.-L.; Ke, Y.-p.; Shi, H.-C.; Yuan, J.-C. Effect of low-nitrogen stress on photosynthesis and chlorophyll fluorescence characteristics of maize cultivars with different low-nitrogen tolerances. J. Integr. Agric. 2019, 18, 1246–1256. [Google Scholar] [CrossRef]
- Akhtar, S.; Mekonnen, T.W.; Mashingaidze, K.; Osthoff, G.; Labuschagne, M.J.H. Heterosis and combining ability of iron, zinc and their bioavailability in maize inbred lines under low nitrogen and optimal environments. Heliyon 2023, 9, e14177. [Google Scholar] [CrossRef]
- Ertiro, B.T.; Das, B.; Kosgei, T.; Tesfaye, A.T.; Labuschagne, M.T.; Worku, M.; Olsen, M.S.; Chaikam, V.; Gowda, M. Relationship between grain yield and quality traits under optimum and low-nitrogen stress environments in tropical maize. Agronomy 2022, 12, 438. [Google Scholar] [CrossRef]
- Abe, A.; Adetimirin, V.O.; Menkir, A.; Moose, S.P.; Olaniyan, A.B. Performance of tropical maize hybrids under conditions of low and optimum levels of nitrogen fertilizer application-grain yield, biomass production and nitrogen accumulation. Maydica 2013, 58, 141–150. [Google Scholar]
- Zhao, M.; Shu, G.; Hu, Y.; Cao, G.; Wang, Y. Pattern and variation in simple sequence repeat (SSR) at different genomic regions and its implications to maize evolution and breeding. BMC Genom. 2023, 24, 136. [Google Scholar] [CrossRef] [PubMed]
- Miyassa, M.A.; Lupini, A.; Mauceri, A.; Morsli, A.; Khelifi, L.; Sunseri, F. Genetic variation and structure of maize populations from Saoura and Gourara oasis in Algerian Sahara. BMC Genom. Data 2018, 19, 51. [Google Scholar] [CrossRef]
- Menkir, A.; Melake-Berhan, A.; The, C.; Ingelbrecht, I.; Adepoju, A. Grouping of tropical mid-altitude maize inbred lines on the basis of yield data and molecular markers. Theor. Appl. Genet. 2004, 108, 1582–1590. [Google Scholar] [CrossRef]
- Wegary, D.; Vivek, B.; Labuschagne, M. Association of parental genetic distance with heterosis and specific combining ability in quality protein maize. Euphytica 2013, 191, 205–216. [Google Scholar] [CrossRef]
- Kamara, M.M.; Ghazy, N.A.; Mansour, E.; Elsharkawy, M.M.; Kheir, A.M.; Ibrahim, K.M. Molecular genetic diversity and line× tester analysis for resistance to late wilt disease and grain yield in maize. Agronomy 2021, 11, 898. [Google Scholar] [CrossRef]
- Reif, J.; Gumpert, F.; Fischer, S.; Melchinger, A. Impact of interpopulation divergence on additive and dominance variance in hybrid populations. Genetics 2007, 176, 1931–1934. [Google Scholar] [CrossRef]
- Adu, G.; Awuku, F.; Amegbor, I.; Haruna, A.; Manigben, K.; Aboyadana, P. Genetic characterization and population structure of maize populations using SSR markers. Ann. Agric. Sci. 2019, 64, 47–54. [Google Scholar] [CrossRef]
- Akinwale, R.; Badu-Apraku, B.; Fakorede, M.; Vroh-Bi, I. Heterotic grouping of tropical early-maturing maize inbred lines based on combining ability in Striga-infested and Striga-free environments and the use of SSR markers for genotyping. Field Crop. Res. 2014, 156, 48–62. [Google Scholar] [CrossRef]
- Labroo, M.R.; Studer, A.J.; Rutkoski, J.E. Heterosis and hybrid crop breeding: A multidisciplinary review. Front. Genet. 2021, 12, 643761. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Araus, J.L.; Shah, T.; Cairns, J.; Mahuku, G.; Bänziger, M.; Torres, J.L.; Sánchez, C.; Yan, J. Molecular characterization of a diverse maize inbred line collection and its potential utilization for stress tolerance improvement. Crop Sci. 2011, 51, 2569–2581. [Google Scholar] [CrossRef]
- Lariepe, A.; Moreau, L.; Laborde, J.; Bauland, C.; Mezmouk, S.; Décousset, L.; Mary-Huard, T.; Fiévet, J.; Gallais, A.; Dubreuil, P. General and specific combining abilities in a maize (Zea mays L.) test-cross hybrid panel: Relative importance of population structure and genetic divergence between parents. Theor. Appl. Genet. 2017, 130, 403–417. [Google Scholar] [CrossRef] [PubMed]
- Owusu, G.A.; Abe, A.; Ribeiro, P.F. Genetic analysis and heterotic grouping of quality protein maize (Zea mays L.) inbred lines and derived hybrids under conditions of low soil nitrogen and drought stress. Euphytica 2023, 219, 29. [Google Scholar] [CrossRef]
- Badu-Apraku, B.; Oyekunle, M.; Fakorede, M.; Vroh, I.; Akinwale, R.O.; Aderounmu, M. Combining ability, heterotic patterns and genetic diversity of extra-early yellow inbreds under contrasting environments. Euphytica 2013, 192, 413–433. [Google Scholar] [CrossRef]
- Betran, F.; Beck, D.; Bänziger, M.; Edmeades, G. Genetic analysis of inbred and hybrid grain yield under stress and nonstress environments in tropical maize. Crop Sci. 2003, 43, 807–817. [Google Scholar] [CrossRef]
- Worku, M.; Bänziger, M.; Friesen, D.; Schulte auf’m Erley, G.; Horst, W.; Vivek, B. Relative importance of general combining ability and specific combining ability among tropical maize (Zea mays L.) inbreds under contrasting nitrogen environments. Maydica 2008, 53, 279–288. [Google Scholar]
- Makinde, S.A.; Badu-Apraku, B.; Ariyo, O.J.; Porbeni, J.B. Combining ability of extra-early maturing pro-vitamin A maize (Zea mays L.) inbred lines and performance of derived hybrids under Striga hermonthica infestation and low soil nitrogen. PLoS ONE 2023, 18, e0280814. [Google Scholar] [CrossRef]



| Parent | IL1 | IL2 | IL3 | IL4 | IL5 | IL6 | IL7 |
|---|---|---|---|---|---|---|---|
| IL1 | 0.00 | ||||||
| IL2 | 0.43 | 0.00 | |||||
| IL3 | 0.71 | 0.78 | 0.00 | ||||
| IL4 | 0.53 | 0.63 | 0.71 | 0.00 | |||
| IL5 | 0.88 | 0.88 | 0.83 | 0.88 | 0.00 | ||
| IL6 | 0.71 | 0.84 | 0.78 | 0.84 | 0.83 | 0.00 | |
| IL7 | 0.65 | 0.72 | 0.72 | 0.79 | 0.74 | 0.65 | 0.00 |
| Source of Variation | df | Days to Silking | Plant Height (cm) | Ear Height (cm) | Ear Length (cm) |
| Nitrogen (N) | 1 | 1334 ** | 174,141 ** | 66,590 ** | 383.5 * |
| Error a | 4 | 12.42 | 3854 | 60.08 | 44.65 |
| Hybrids (H) | 20 | 50.37 ** | 1705 ** | 978.7 ** | 18.50 ** |
| GCA | 6 | 58.91 ** | 1755 ** | 1374 ** | 34.64 ** |
| SCA | 14 | 46.71 ** | 1684 ** | 809.3 ** | 11.58 ** |
| H×N | 20 | 6.31 * | 299.9 * | 511.9 ** | 10.52 ** |
| GCA×N | 6 | 4.15 | 388.8 * | 454.3 ** | 8.71 ** |
| SCA×N | 14 | 7.24 * | 261.8 | 536.6 ** | 11.29 ** |
| Error b | 80 | 3.63 | 156.4 | 90.85 | 0.75 |
| GCA/SCA | 1.26 | 1.04 | 1.70 | 2.99 | |
| GCA×N/SCA×N | 0.57 | 1.48 | 0.85 | 0.77 | |
| SOV | df | Number of Rows per Ear | Number of Kernels per Row | Thousand Kernel Weight (g) | Grain Yield (t/ha) |
| Nitrogen (N) | 1 | 118.1 ** | 1551 ** | 211,396 ** | 141.6 ** |
| Error a | 4 | 1.21 | 1.27 | 200.3 | 1.06 |
| Hybrids (H) | 20 | 6.89 ** | 114.0 ** | 5305 ** | 13.03 ** |
| GCA | 6 | 11.30 ** | 230.4 ** | 9144 ** | 33.48 ** |
| SCA | 14 | 5.00 ** | 64.09 ** | 3660 ** | 4.27 ** |
| H×N | 20 | 1.28 * | 35.12 ** | 769.2 ** | 1.72 ** |
| GCA×N | 6 | 1.42 | 42.69 * | 976.5 ** | 3.21 ** |
| SCA×N | 14 | 1.22 * | 31.88 * | 680.4 ** | 1.08 ** |
| Error b | 80 | 0.66 | 14.13 | 149.1 | 0.15 |
| GCA/SCA | 2.26 | 3.60 | 2.50 | 7.84 | |
| GCA×N/SCA×N | 1.16 | 1.34 | 1.44 | 2.97 |
| Line | Days to Silking | Plant Height (cm) | Ear Height (cm) | Ear Length (cm) | ||||
| Low N | Recommended N | Low N | Recommended N | Low N | Recommended N | Low N | Recommended N | |
| IL1 | −1.22 * | −2.10 ** | 6.66 | 8.86 ** | 7.58 ** | 6.74 ** | 1.22 ** | 1.78 ** |
| IL2 | −0.89 | 0.44 | 11.53 ** | 9.60 ** | 5.45 * | 16.87 ** | 0.80 ** | 0.84 ** |
| IL3 | −2.23 ** | −1.70 ** | −1.76 | 7.10 * | −7.75 ** | −5.89 * | 1.09 ** | 0.22 |
| IL4 | 0.18 | −0.16 | −4.00 | −7.49 ** | 2.65 | −3.01 | −1.10 ** | 0.51 * |
| IL5 | 1.58 ** | 0.97 * | −1.74 | 2.94 | −6.69 ** | −1.67 | −1.02 ** | −2.08 ** |
| IL6 | 0.85 | 0.97 * | 1.33 | −12.09 ** | 5.11 * | −7.93 ** | 0.48 * | −0.69 ** |
| IL7 | 1.71 ** | 1.57 ** | −12.03 ** | −8.92 ** | −6.35 ** | −5.13 * | −1.48 ** | −0.58 * |
| LSD (0.05) gi | 0.95 | 0.89 | 6.67 | 5.34 | 4.60 | 4.61 | 0.39 | 0.45 |
| LSD (0.01) gi | 1.27 | 1.19 | 8.93 | 7.14 | 6.15 | 6.17 | 0.52 | 0.60 |
| Inbred Line | No. of Rows per Ear | No. of Kernels per Row | 1000-Kernel Weight (g) | Grain Yield (t/ha) | ||||
| Low N | Recommended N | Low N | Recommended N | Low N | Recommended N | Low N | Recommended N | |
| IL1 | 0.54 * | 0.89 ** | 2.75 ** | 4.80 ** | 10.63 ** | 17.52 ** | 0.95 ** | 1.26 ** |
| IL2 | 0.68 ** | 0.22 | 3.82 ** | 1.67 * | 32.63 ** | 15.52 ** | 1.61 ** | 1.07 ** |
| IL3 | 0.34 | 0.35 | 2.22 * | 0.00 | 9.16 ** | 14.86 ** | 0.73 ** | 0.87 ** |
| IL4 | −1.39 ** | −0.85 ** | −0.91 | 0.07 | −17.77 ** | −13.14 ** | −1.36 ** | −0.68 ** |
| IL5 | −0.06 | −0.71 ** | −1.58 | −1.00 | −0.50 | −14.81 ** | −0.52 ** | −1.23 ** |
| IL6 | −0.26 | 0.02 | −0.25 | −2.93 ** | −2.84 | −2.14 | 0.07 | −0.63 ** |
| IL7 | 0.14 | 0.09 | −6.05 ** | −2.60 ** | −31.30 ** | −17.81 ** | −1.48 ** | −0.65 ** |
| LSD (0.05) gi | 0.41 | 0.37 | 2.03 | 1.57 | 6.37 | 5.39 | 0.18 | 0.19 |
| LSD (0.01) gi | 0.55 | 0.50 | 2.72 | 2.10 | 8.52 | 7.21 | 0.24 | 0.26 |
| Cross | Days to Silking | Plant Height (cm) | Ear Height (cm) | Ear Length (cm) | ||||
| Low N | Recommended N | Low N | Recommended N | Low N | Recommended N | Low N | Recommended N | |
| IL1×IL2 | 2.98 ** | 2.02 * | −3.14 | 13.68 * | 21.73 ** | −7.50 | 1.54 ** | −0.44 |
| IL1×IL3 | 0.31 | 3.82 ** | 12.82 | 3.40 | 19.93 ** | 21.26 ** | −1.04 ** | 1.59 ** |
| IL1×IL4 | −2.42 * | −1.38 | 15.06 * | 1.00 | −1.13 | 6.72 | 2.51 ** | −1.74 ** |
| IL1×IL5 | 1.84 | −0.18 | 27.12 ** | 20.56 ** | −18.47 ** | 11.72 * | −1.50 ** | −0.55 |
| IL1×IL6 | −0.09 | 2.16 * | −22.28 ** | −4.40 | −5.60 | −15.36 ** | −1.40 ** | 1.79 ** |
| IL1×IL7 | −2.62 ** | −6.44 ** | −29.58 ** | −34.24 ** | −16.47 ** | −16.83 ** | −0.10 | −0.65 |
| IL2×IL3 | −0.02 | −3.04 ** | −1.05 | −12.33 * | −17.93 ** | −16.20 ** | −0.46 | −0.98 * |
| IL2×IL4 | 0.91 | 0.09 | 5.19 | −5.74 | −7.00 | 4.25 | −1.84 ** | −4.08 ** |
| IL2×IL5 | 0.51 | 1.62 | 1.92 | 11.33 * | −8.33 | 4.92 | 0.19 | 1.79 ** |
| IL2×IL6 | −6.09 ** | −4.04 ** | −12.14 | −5.14 | 14.20 ** | 16.50 ** | 0.82 * | 2.02 ** |
| IL2×IL7 | 1.71 | 3.36 ** | 9.22 | −1.81 | −2.67 | −1.96 | −0.25 | 1.69 ** |
| IL3×IL4 | −2.09 * | −2.11 * | −13.52 * | −9.24 | −1.80 | 15.61 ** | 0.11 | 2.15 ** |
| IL3×IL5 | −2.82 ** | −1.91 * | 4.88 | 12.32 * | 10.20 * | −12.39 ** | 1.70 ** | −2.12 ** |
| IL3×IL6 | 4.24 ** | 2.42 ** | 12.15 | 5.16 | −12.27 ** | −6.07 | 2.00 ** | 0.08 |
| IL3×IL7 | 0.38 | 0.82 | −15.29 * | 0.68 | 1.87 | −2.20 | −2.30 ** | −0.72 |
| IL4×IL5 | 1.11 | 0.22 | −26.88 ** | −13.08 * | −5.53 | −6.54 | −1.51 ** | 1.42 ** |
| IL4×IL6 | 3.18 ** | 2.56 ** | 10.39 | 6.95 | 6.67 | −5.28 | 0.38 | −0.21 |
| IL4×IL7 | −0.69 | 0.62 | 9.75 | 20.11 ** | 8.80 | −14.75 ** | 0.35 | 2.47 ** |
| IL5×IL6 | −1.56 | −2.24 * | −10.54 | −24.48 ** | 5.33 | −11.62 * | −1.49 ** | −0.72 |
| IL5×IL7 | 0.91 | 2.49 ** | 3.48 | −6.66 | 16.80 ** | 13.92 ** | 2.61 ** | 0.18 |
| IL6×IL7 | 0.31 | −0.84 | 22.42 ** | 21.91 ** | −8.33 | 21.84 ** | −0.30 | −2.97 ** |
| LSD 5% (sij) | 1.87 | 1.76 | 13.16 | 10.52 | 9.07 | 9.09 | 0.76 | 0.88 |
| LSD 1% (sij) | 2.50 | 2.35 | 17.60 | 14.08 | 12.13 | 12.17 | 1.02 | 1.18 |
| Cross | No. of Rows per Ear | No. of Kernels per Row | 1000-Kernel Weight (g) | Grain Yield (t/ha) | ||||
| Low N | Recommended N | Low-N | Recommended N | Low N | Recommended N | Low-N | Recommended N | |
| IL1×IL2 | 1.18 ** | 1.36 ** | 0.44 | 0.87 | 13.82 * | 13.78 * | 0.08 | 0.48 * |
| IL1×IL3 | 1.18 ** | 0.89 * | −0.29 | 1.87 | −26.04 ** | −15.56 ** | 0.56 ** | 0.67 ** |
| IL1×IL4 | 0.58 | 0.09 | −1.16 | 0.13 | −20.44 ** | 12.44 * | 0.64 ** | 0.00 |
| IL1×IL5 | −1.42 ** | −0.38 | 2.51 | −0.80 | 20.29 ** | 24.11 ** | −1.55 ** | −0.75 ** |
| IL1×IL6 | −0.56 | −0.44 | 2.18 | 2.13 | 19.29 ** | 11.44 * | 0.77 ** | 0.56 ** |
| IL1×IL7 | −0.96 * | −1.51 ** | −3.69 | −4.20 ** | −6.91 | −46.22 ** | −0.49 ** | −0.96 ** |
| IL2×IL3 | −0.62 | −1.11 ** | −3.02 | 0.00 | −1.38 | −3.56 | 0.13 | −1.10 ** |
| IL2×IL4 | −0.22 | 1.42 ** | −7.22 ** | −4.40 ** | 18.89 ** | 14.44 ** | −1.13 ** | −0.84 ** |
| IL2×IL5 | 0.44 | −1.38 ** | 2.11 | 0.00 | −38.38 ** | −23.89 ** | 0.66 ** | 0.16 |
| IL2×IL6 | −1.36 ** | −0.78 * | 2.44 | 4.60 ** | 0.62 | 0.11 | 0.79 ** | 0.22 |
| IL2×IL7 | 0.58 | 0.49 | 5.24 * | −1.07 | 6.42 | −0.89 | −0.52 ** | 1.08 ** |
| IL3×IL4 | 0.11 | −0.71 | −0.62 | 2.60 | 39.02 ** | 35.11 ** | −0.26 | 1.04 ** |
| IL3×IL5 | 0.44 | 0.82 * | 4.04 * | 4.00 * | 15.09 * | 6.78 | 1.16 ** | 0.85 ** |
| IL3×IL6 | 0.31 | 0.42 | −1.29 | −6.07 ** | 7.42 | −2.56 | −0.37 * | −0.09 |
| IL3×IL7 | −1.42 ** | −0.31 | 1.18 | −2.40 | −34.11 ** | −20.22 ** | −1.21 ** | −1.37 ** |
| IL4×IL5 | −0.16 | −0.31 | 1.18 | 0.60 | −27.98 ** | −36.89 ** | −0.39 * | −0.65 ** |
| IL4×IL6 | 0.04 | −0.04 | 5.18 * | −4.13 * | −12.31 | −42.89 ** | −0.18 | −0.69 ** |
| IL4×IL7 | −0.36 | −0.44 | 2.64 | 5.20 ** | 2.82 | 17.78 ** | 1.33 ** | 1.13 ** |
| IL5×IL6 | 0.04 | 0.16 | −6.49 ** | −1.40 | −7.91 | 7.11 | −0.88 ** | 0.13 |
| IL5×IL7 | 0.64 | 1.09 ** | −3.36 | −2.40 | 38.89 ** | 22.78 ** | 1.01 ** | 0.26 |
| IL6×IL7 | 1.51 ** | 0.69 | −2.02 | 4.87 ** | −7.11 | 26.78 ** | −0.12 | −0.14 |
| LSD 5% (sij) | 0.80 | 0.74 | 4.01 | 3.09 | 12.56 | 10.63 | 0.35 | 0.38 |
| LSD 1% (sij) | 1.08 | 0.99 | 5.37 | 4.14 | 16.80 | 14.22 | 0.47 | 0.51 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamara, M.M.; Mansour, E.; Khalaf, A.E.A.; Eid, M.A.M.; Hassanin, A.A.; Abdelghany, A.M.; Kheir, A.M.S.; Galal, A.A.; Behiry, S.I.; Silvar, C.; et al. Molecular Diversity and Combining Ability in Newly Developed Maize Inbred Lines under Low-Nitrogen Conditions. Life 2024, 14, 641. https://doi.org/10.3390/life14050641
Kamara MM, Mansour E, Khalaf AEA, Eid MAM, Hassanin AA, Abdelghany AM, Kheir AMS, Galal AA, Behiry SI, Silvar C, et al. Molecular Diversity and Combining Ability in Newly Developed Maize Inbred Lines under Low-Nitrogen Conditions. Life. 2024; 14(5):641. https://doi.org/10.3390/life14050641
Chicago/Turabian StyleKamara, Mohamed M., Elsayed Mansour, Ahmed E. A. Khalaf, Mohamed A. M. Eid, Abdallah A. Hassanin, Ahmed M. Abdelghany, Ahmed M. S. Kheir, Ahmed A. Galal, Said I. Behiry, Cristina Silvar, and et al. 2024. "Molecular Diversity and Combining Ability in Newly Developed Maize Inbred Lines under Low-Nitrogen Conditions" Life 14, no. 5: 641. https://doi.org/10.3390/life14050641













