PIGN-Related Disease in Two Lithuanian Families: A Report of Two Novel Pathogenic Variants, Molecular and Clinical Characterisation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical Evaluation of the Individuals
2.2. DNA Extraction
2.3. Next-Generation Sequencing
2.4. Sanger Sequencing
2.5. Western Blot
2.6. Post-Sequencing Data Analysis
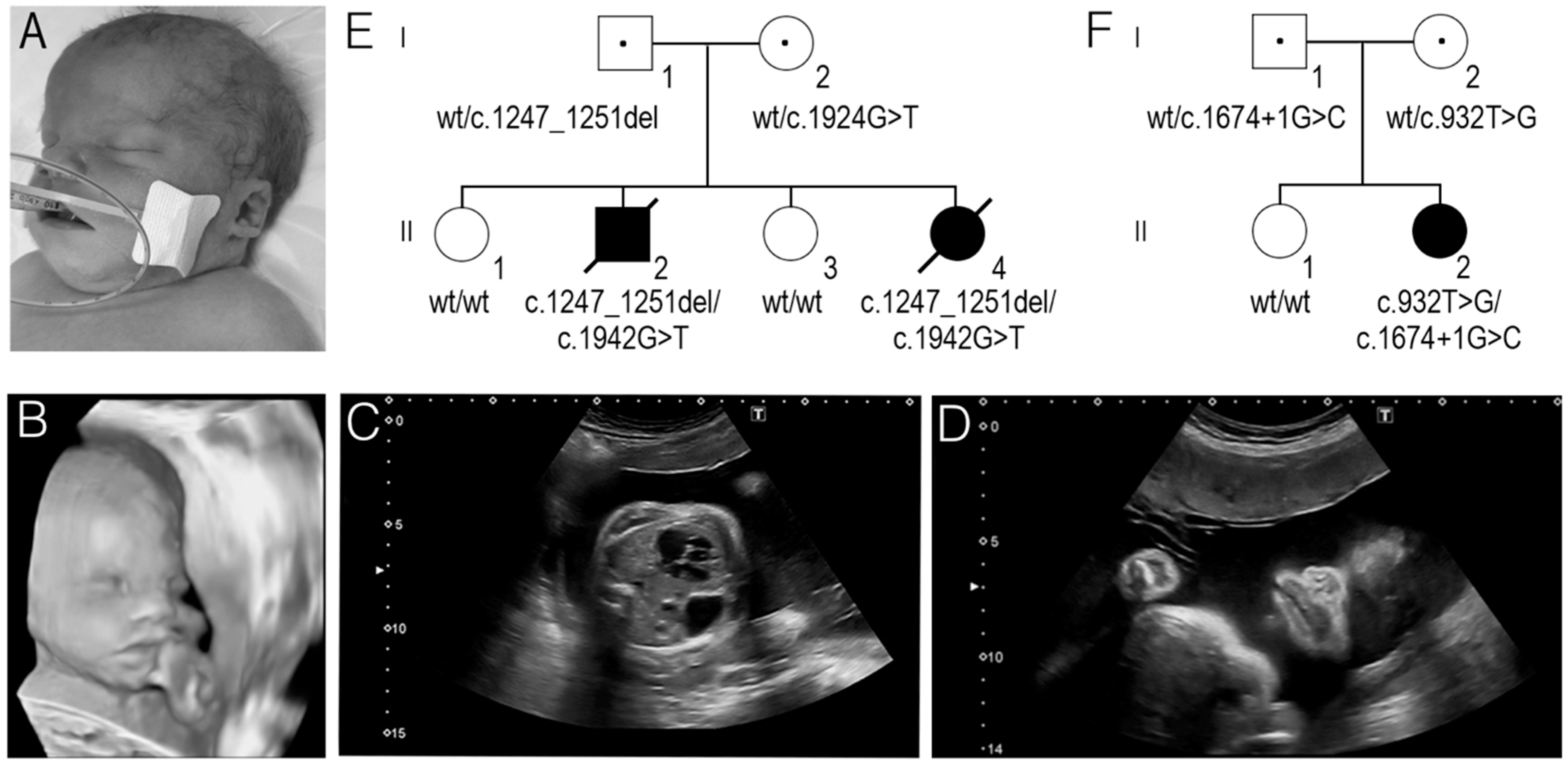
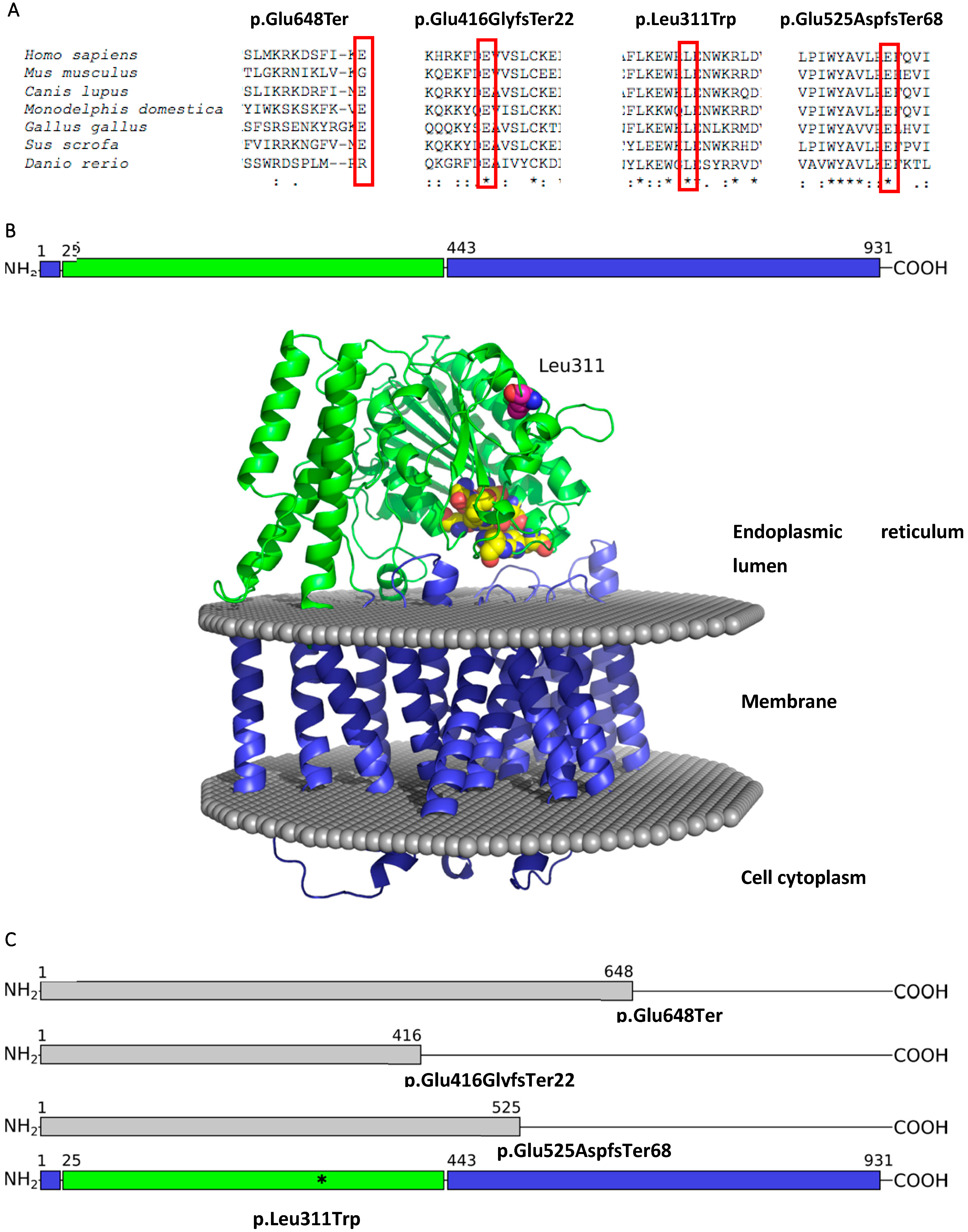
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACMG | American College of Human Genetics and Genomics; |
| CDH | Congenital diaphragmatic hernia; |
| EtNP | Ethanolamine phosphate; |
| FS | Fryns syndrome; |
| gDNA | Genomic DNA; |
| GPI | Glycosylphosphatidylinositol; |
| IGV | Integrative Genomics Viewer; |
| MCAHS | Multiple congenital anomalies-hypotonia-seizures syndrome; |
| MCAHS1 | Multiple congenital anomalies-hypotonia-seizures syndrome 1; |
| mRNA | Messenger RNA; |
| NGS | Next-generation sequencing; |
| PCR | Polymerase chain reaction; |
| PIGA | Phosphatidylinositol glycan biosynthesis class A; |
| PIGN | Phosphatidylinositol glycan biosynthesis class N; |
| PIGQ | Phosphatidylinositol glycan biosynthesis class Q; |
| PIGT | Phosphatidylinositol glycan biosynthesis class T; |
| VCF | Variant calling format; |
| WES | Whole exome sequencing. |
References
- Knaus, A.; Pantel, J.T.; Pendziwiat, M.; Hajjir, N.; Zhao, M.; Hsieh, T.-C.; Schubach, M.; Gurovich, Y.; Fleischer, N.; Jäger, M.; et al. Characterization of glycosylphosphatidylinositol biosynthesis defects by clinical features, flow cytometry, and automated image analysis. Genome Med. 2018, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Jiao, X.; Xue, J.; Gong, P.; Bao, X.; Wu, Y.; Zhang, Y.; Jiang, Y.; Yang, Z. Analyzing clinical and genetic characteristics of a cohort with multiple congenital anomalies-hypotonia-seizures syndrome (MCAHS). Orphanet J. Rare Dis. 2020, 15, 78. [Google Scholar] [CrossRef] [PubMed]
- Maydan, G.; Noyman, I.; Har-Zahav, A.; Ben Neriah, Z.; Pasmanik-Chor, M.; Yeheskel, A.; Albin-Kaplanski, A.; Maya, I.; Magal, N.; Birk, E.; et al. Multiple congenital anomalies-hypotonia-seizures syndrome is caused by a mutation in PIGN. J. Med. Genet. 2011, 48, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Stenson, P.D.; Ball, E.V.; Mort, M.; Phillips, A.D.; Shiel, J.A.; Thomas, N.S.; Abeysinghe, S.; Krawczak, M.; Cooper, D.N. Human Gene Mutation Database (HGMD®): 2003 update. Hum. Mutat. 2003, 21, 577–581. [Google Scholar] [CrossRef] [PubMed]
- Brady, P.; Moerman, P.; De Catte, L.; Deprest, J.; Devriendt, K.; Vermeesch, J.R. Exome sequencing identifies a recessive PIGN splice site mutation as a cause of syndromic Congenital Diaphragmatic Hernia. Eur. J. Med. Genet. 2014, 57, 487–493. [Google Scholar] [CrossRef]
- McInerney-Leo, A.M.; Harris, J.E.; Gattas, M.; Peach, E.E.; Sinnott, S.; Dudding-Byth, T.; Rajagopalan, S.; Barnett, C.P.; Anderson, L.K.; Wheeler, L.; et al. Fryns Syndrome Associated with Recessive Mutations in PIGN in two Separate Families. Hum. Mutat. 2016, 37, 695–702. [Google Scholar] [CrossRef]
- Fryns, J.P.; Moerman, F.; Goddeeris, P.; Bossuyt, C.; Van den Berghe, H. A new lethal syndrome with cloudy corneae, diaphragmatic defects and distal limb deformities. Hum. Genet. 1979, 50, 65–70. [Google Scholar] [CrossRef]
- Lin, A.E.; Pober, B.R.; Mullen, M.P.; Slavotinek, A.M. Cardiovascular malformations in Fryns syndrome: Is there a pathogenic role for neural crest cells? Am. J. Med. Genet. Part A 2005, 139A, 186–193. [Google Scholar] [CrossRef]
- Bayat, A.; de Valles-Ibáñez, G.; Pendziwiat, M.; Knaus, A.; Alt, K.; Biamino, E.; Bley, A.; Calvert, S.; Carney, P.; Caro-Llopis, A.; et al. PIGN encephalopathy: Characterizing the epileptology. Epilepsia 2022, 63, 974–991. [Google Scholar] [CrossRef]
- Tian, M.; Chen, J.; Li, J.; Pan, H.; Lei, W.; Shu, X. Damaging novel mutations in PIGN cause developmental epileptic-dyskinetic encephalopathy: A case report. BMC Pediatr. 2022, 22, 222. [Google Scholar] [CrossRef]
- Thiffault, I.; Zuccarelli, B.; Welsh, H.; Yuan, X.; Farrow, E.; Zellmer, L.; Miller, N.; Soden, S.; Abdelmoity, A.; Brodsky, R.A.; et al. Hypotonia and intellectual disability without dysmorphic features in a patient with PIGN-related disease. BMC Med. Genet. 2017, 18, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Javadi, A.; Shamaei, M.; Ziazi, L.M.; Pourabdollah, M.; Dorudinia, A.; Seyedmehdi, S.M.; Karimi, S. Qualification Study of Two Genomic DNA Extraction Methods in Different Clinical Samples. Tanaffos 2014, 13, 41–47. [Google Scholar] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, J.T.; Thorvaldsdóttir, H.; Wenger, A.M.; Zehir, A.; Mesirov, J.P. Variant Review with the Integrative Genomics Viewer. Cancer Res. 2017, 77, e31–e34. [Google Scholar] [CrossRef] [Green Version]
- Desvignes, J.-P.; Bartoli, M.; Delague, V.; Krahn, M.; Miltgen, M.; Béroud, C.; Salgado, D. VarAFT: A variant annotation and filtration system for human next generation sequencing data. Nucleic Acids Res. 2018, 46, W545–W553. [Google Scholar] [CrossRef]
- Jaganathan, K.; Panagiotopoulou, S.K.; McRae, J.F.; Darbandi, S.F.; Knowles, D.; Li, Y.I.; Kosmicki, J.A.; Arbelaez, J.; Cui, W.; Schwartz, G.B.; et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell 2019, 176, 535–548.e24. [Google Scholar] [CrossRef] [Green Version]
- Hubbard, T.; Barker, D.; Birney, E.; Cameron, G.; Chen, Y.; Clark, L.; Cox, T.; Cuff, J.; Curwen, V.; Down, T.; et al. The Ensembl genome database project. Nucleic Acids Res. 2002, 30, 38–41. [Google Scholar] [CrossRef] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Gasteiger, E. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bateman, A. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Šali, A.; Blundell, T.L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Dapkūnas, J.; Margelevičius, M. The COMER web server for protein analysis by homology. 2022; submitted. [Google Scholar]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; López, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [Green Version]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator: Figure 1. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [Green Version]
- Lomize, M.A.; Pogozheva, I.D.; Joo, H.; Mosberg, H.I.; Lomize, A.L. OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 2012, 40, D370–D376. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Riley, G.R.; Jang, W.; Rubinstein, W.S.; Church, D.M.; Maglott, D.R. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014, 42, D980–D985. [Google Scholar] [CrossRef] [Green Version]
- Stenson, P.D.; Mort, M.; Ball, E.V.; Evans, K.; Hayden, M.; Heywood, S.; Hussain, M.; Phillips, A.D.; Cooper, D.N. The Human Gene Mutation Database: Towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 2017, 136, 665–677. [Google Scholar] [CrossRef]
- Jezela-Stanek, A.; Ciara, E.; Piekutowska-Abramczuk, D.; Trubicka, J.; Jurkiewicz, E.; Rokicki, D.; Mierzewska, H.; Spychalska, J.; Uhrynowska, M.; Szwarc-Bronikowska, M.; et al. Congenital disorder of glycosylphosphatidylinositol (GPI)-anchor biosynthesis—The phenotype of two patients with novel mutations in the PIGN and PGAP2 genes. Eur. J. Paediatr. Neurol. 2016, 20, 462–473. [Google Scholar] [CrossRef]
- Pronicka, E.; Piekutowska-Abramczuk, D.; Ciara, E.; Trubicka, J.; Rokicki, D.; Karkucińska-Więckowska, A.; Pajdowska, M.; Jurkiewicz, E.; Halat, P.; Kosińska, J.; et al. New perspective in diagnostics of mitochondrial disorders: Two years’ experience with whole-exome sequencing at a national paediatric centre. J. Transl. Med. 2016, 14, 174. [Google Scholar] [CrossRef] [Green Version]
- Pagnamenta, A.T.; the DDD study; Murakami, Y.; Taylor, J.M.; Anzilotti, C.; Howard, M.F.; Miller, V.; Johnson, D.S.; Tadros, S.; Mansour, S.; et al. Analysis of exome data for 4293 trios suggests GPI-anchor biogenesis defects are a rare cause of developmental disorders. Eur. J. Hum. Genet. 2017, 25, 669–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleming, L.; Lemmon, M.; Beck, N.; Johnson, M.; Mu, W.; Murdock, D.; Bodurtha, J.; Hoover-Fong, J.; Cohn, R.; Bosemani, T.; et al. Genotype-phenotype correlation of congenital anomalies in multiple congenital anomalies hypotonia seizures syndrome (MCAHS1)/PIGN-related epilepsy. Am. J. Med. Genet. Part A 2016, 170, 77–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakagawa, T.; Taniguchi-Ikeda, M.; Murakami, Y.; Nakamura, S.; Motooka, D.; Emoto, T.; Satake, W.; Nishiyama, M.; Toyoshima, D.; Morisada, N.; et al. A novel PIGN mutation and prenatal diagnosis of inherited glycosylphosphatidylinositol deficiency. Am. J. Med. Genet. Part A 2016, 170, 183–188. [Google Scholar] [CrossRef] [PubMed]
- De Giorgis, V.; Paoletti, M.; Varesio, C.; Gana, S.; Rognone, E.; Dallavalle, G.; Papalia, G.; Pichiecchio, A. Novel insights into the clinico-radiological spectrum of phenotypes associated to PIGN mutations. Eur. J. Paediatr. Neurol. 2021, 33, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Alessandri, J.-L.; Gordon, C.T.; Jacquemont, M.-L.; Gruchy, N.; Ajeawung, N.F.; Benoist, G.; Oufadem, M.; Chebil, A.; Duffourd, Y.; Dumont, C.; et al. Recessive loss of function PIGN alleles, including an intragenic deletion with founder effect in la Réunion Island, in patients with Fryns syndrome. Eur. J. Hum. Genet. 2018, 26, 340–349. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Eisenhaber, B.; Maurer-Stroh, S.; Novatchkova, M.; Schneider, G.; Eisenhaber, F. Enzymes and auxiliary factors for GPI lipid anchor biosynthesis and post-translational transfer to proteins. BioEssays 2003, 25, 367–385. [Google Scholar] [CrossRef]
- Kinoshita, T. Biosynthesis and deficiencies of glycosylphosphatidylinositol. Proc. Japan Acad. Ser. B 2014, 90, 130–143. [Google Scholar] [CrossRef]
- McKean, D.M.; Niswander, L. Defects in GPI biosynthesis perturb Cripto signaling during forebrain development in two new mouse models of holoprosencephaly. Biol. Open 2012, 1, 874–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, G.-H.; Fujita, M.; Takaoka, K.; Murakami, Y.; Fujihara, Y.; Kanzawa, N.; Murakami, K.-I.; Kajikawa, E.; Takada, Y.; Saito, K.; et al. A GPI processing phospholipase A2, PGAP6, modulates Nodal signaling in embryos by shedding CRIPTO. J. Cell Biol. 2016, 215, 705–718. [Google Scholar] [CrossRef] [Green Version]
- Baranova, A.; Cao, H.; Zhang, F. Unraveling Risk Genes of COVID-19 by Multi-Omics Integrative Analyses. Front. Med. 2021, 8, 738687. [Google Scholar] [CrossRef] [PubMed]
- Teye, E.K.; Lu, S.; Chen, F.; Yang, W.; Abraham, T.; Stairs, D.B.; Wang, H.-G.; Yochum, G.S.; Brodsky, R.A.; Pu, J.J. PIGN spatiotemporally regulates the spindle assembly checkpoint proteins in leukemia transformation and progression. Sci. Rep. 2021, 11, 19022. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y.; Jedrzejas, M.J. Conserved core structure and active site residues in alkaline phosphatase superfamily enzymes. Proteins Struct. Funct. Genet. 2001, 45, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Tisserant, E.; Vitobello, A.; Callegarin, D.; Verdez, S.; Bruel, A.; Glele, L.S.A.; Sorlin, A.; Viora-Dupont, E.; Konyukh, M.; Marle, N.; et al. Copy number variants calling from WES data through eXome hidden Markov model (XHMM) identifies additional 2.5% pathogenic genomic imbalances smaller than 30 kb undetected by array-CGH. Ann. Hum. Genet. 2022, 86, 171–180. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siavrienė, E.; Maldžienė, Ž.; Mikštienė, V.; Petraitytė, G.; Rančelis, T.; Dapkūnas, J.; Burnytė, B.; Benušienė, E.; Sasnauskienė, A.; Grikinienė, J.; et al. PIGN-Related Disease in Two Lithuanian Families: A Report of Two Novel Pathogenic Variants, Molecular and Clinical Characterisation. Medicina 2022, 58, 1526. https://doi.org/10.3390/medicina58111526
Siavrienė E, Maldžienė Ž, Mikštienė V, Petraitytė G, Rančelis T, Dapkūnas J, Burnytė B, Benušienė E, Sasnauskienė A, Grikinienė J, et al. PIGN-Related Disease in Two Lithuanian Families: A Report of Two Novel Pathogenic Variants, Molecular and Clinical Characterisation. Medicina. 2022; 58(11):1526. https://doi.org/10.3390/medicina58111526
Chicago/Turabian StyleSiavrienė, Evelina, Živilė Maldžienė, Violeta Mikštienė, Gunda Petraitytė, Tautvydas Rančelis, Justas Dapkūnas, Birutė Burnytė, Eglė Benušienė, Aušra Sasnauskienė, Jurgita Grikinienė, and et al. 2022. "PIGN-Related Disease in Two Lithuanian Families: A Report of Two Novel Pathogenic Variants, Molecular and Clinical Characterisation" Medicina 58, no. 11: 1526. https://doi.org/10.3390/medicina58111526
APA StyleSiavrienė, E., Maldžienė, Ž., Mikštienė, V., Petraitytė, G., Rančelis, T., Dapkūnas, J., Burnytė, B., Benušienė, E., Sasnauskienė, A., Grikinienė, J., Griškevičiūtė, E., Utkus, A., & Preikšaitienė, E. (2022). PIGN-Related Disease in Two Lithuanian Families: A Report of Two Novel Pathogenic Variants, Molecular and Clinical Characterisation. Medicina, 58(11), 1526. https://doi.org/10.3390/medicina58111526





