Untargeted Metabolomics Analysis by UHPLC-MS/MS of Soybean Plant in a Compatible Response to Phakopsora pachyrhizi Infection
Abstract
:1. Introduction
2. Results
2.1. Visual Observation of Symptoms
2.2. UHPLC-ESI(+)-MS/MS Analysis
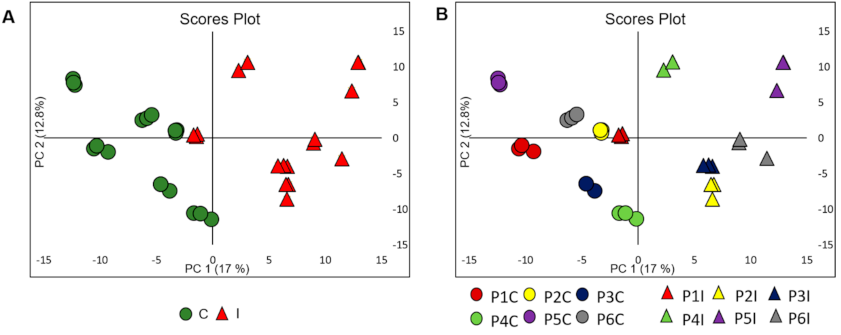
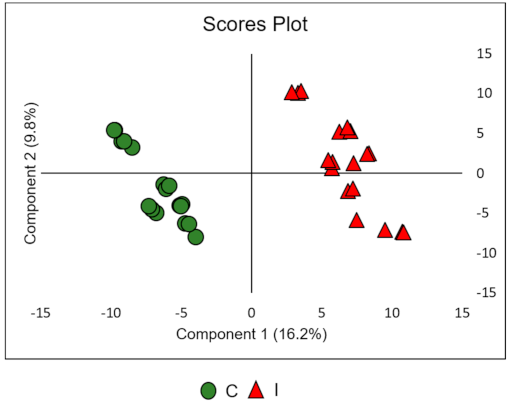
2.3. Data Processing and Chemometric Analysis
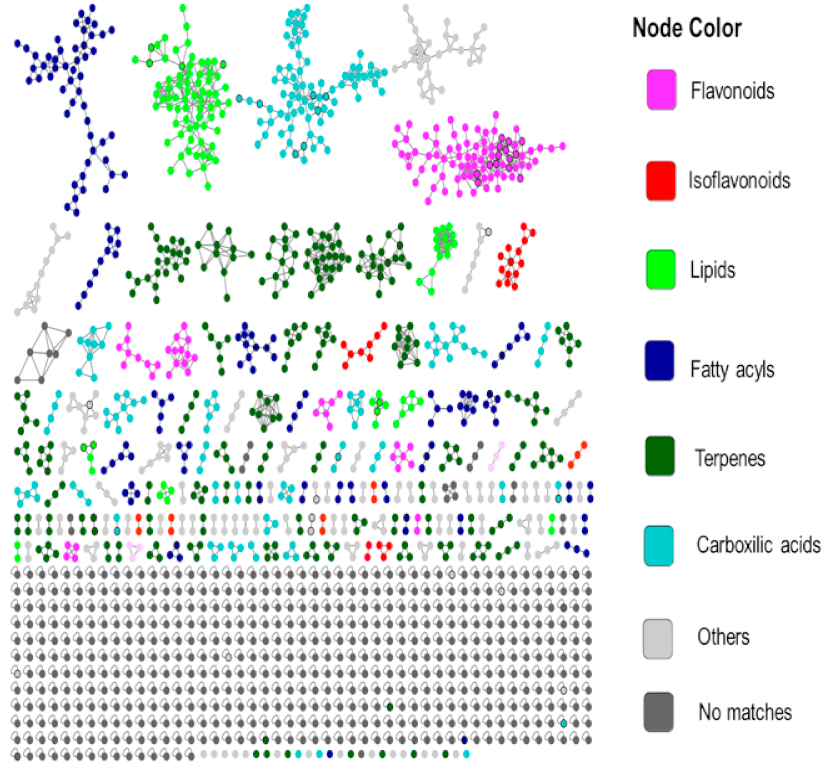
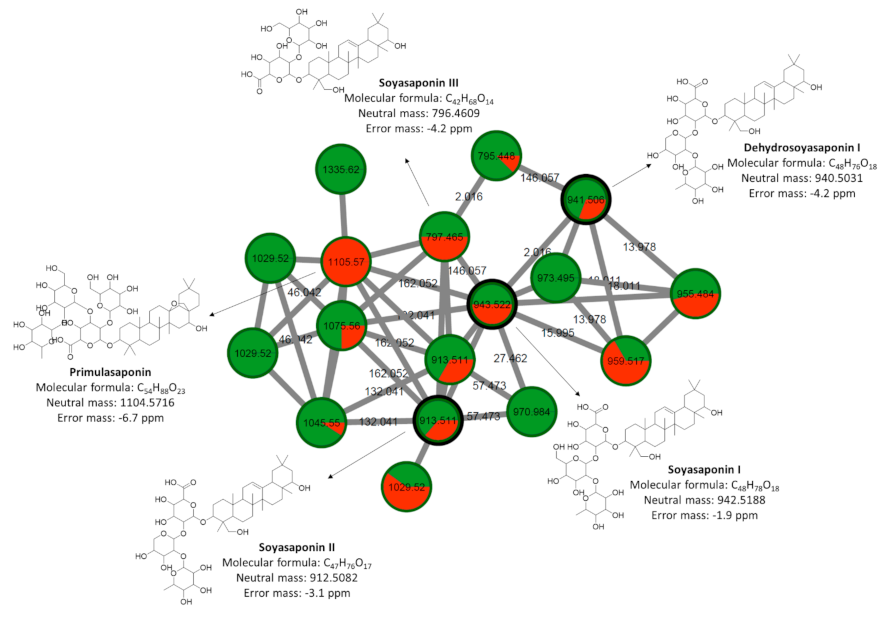
2.4. Chemical Classification and Identification of Metabolites by the GNPS Platform
3. Discussion
4. Materials and Methods
4.1. Plant Preparation
4.2. Preparation and Inoculation of the P. pachyrhizi
4.3. Metabolites Extraction
4.4. UHPLC-ESI-MS/MS Analysis
4.5. Data Preprocessing and Data Analysis
4.6. Classical Molecular Networking Workflow Description
4.7. MolNetEnhancer Workflow Description for Chemical Class Annotation of Molecular Networks
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bratton, M.R.; Martin, E.C.; Elliott, S.; Rhodes, L.V.; Collins-Burow, B.M.; McLachlan, J.A.; Wiese, T.E.; Boue, S.M.; Burow, M.E. Glyceollin, a novel regulator of mTOR/p70S6 in estrogen receptor positive breast cancer. J. Steroid Biochem. Mol. Biol. 2015, 150, 17–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, J.; Nelson, E.; Tilman, D.; Polasky, S.; Tiffany, D. Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels. Proc. Natl. Acad. Sci. USA 2006, 103, 11206–11210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, T.W.; Bradley, C.A.; Sisson, A.J.; Byamukama, E.; Chilvers, M.I.; Coker, C.M.; Collins, A.A.; Damicone, J.P.; Dorrance, A.E.; Dufault, N.S.; et al. Soybean yield loss estimates due to diseases in the United States and Ontario, Canada, from 2010 to 2014. Plant Health Prog. 2017, 18, 19–27. [Google Scholar] [CrossRef] [Green Version]
- Hossain, M.Z.; Ishiga, Y.; Yamanaka, N.; Ogiso-Tanaka, E.; Yamaoka, Y. Soybean leaves transcriptomic data dissects the phenylpropanoid pathway genes as a defence response against Phakopsora pachyrhizi. Plant Physiol. Biochem. 2018, 132, 424–433. [Google Scholar] [CrossRef]
- Qi, M.; Grayczyk, J.P.; Seitz, J.M.; Lee, Y.; Link, T.I.; Choi, D.; Pedley, K.F.; Voegele, R.T.; Baum, T.J.; Whitham, S.A. Suppression or activation of immune responses by predicted secreted proteins of the soybean rust pathogen Phakopsora pachyrhizi. Mol. Plant Microbe Interact. 2018, 31, 163–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Twizeyimana, M.; Hartman, G.L. Sensitivity of Phakopsora pachyrhizi isolates to fungicides and reduction of fungal infection based on fungicide and timing of application. Plant Dis. 2017, 101, 121–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godoy, C.V. Risk and management of fungicide resistance in the Asian soybean rust fungus Phakopsora pachyrhizi. In Fungicide Resistance in Crop Protection: Risk and Management; National Soybean Research Center: Londrina, Brazil, 2011; pp. 87–95. ISBN 9781845939052. [Google Scholar]
- Langenbach, C.; Campe, R.; Beyer, S.F.; Mueller, A.N.; Conrath, U. Fighting Asian soybean rust. Front. Plant Sci. 2016, 7, 797. [Google Scholar] [CrossRef] [Green Version]
- Childs, S.P.; King, Z.R.; Walker, D.R.; Harris, D.K.; Pedley, K.F.; Buck, J.W.; Boerma, H.R.; Li, Z. Discovery of a seventh Rpp soybean rust resistance locus in soybean accession PI 605823. Theor. Appl. Genet. 2018, 131, 27–41. [Google Scholar] [CrossRef]
- Yamanaka, N.; Hossain, M.M. Pyramiding three rust-resistance genes confers a high level of resistance in soybean (Glycine max). Plant Breed. 2019, 138, 686–695. [Google Scholar] [CrossRef]
- Murithi, H.M.; Beed, F.; Tukamuhabwa, P.; Thomma, B.P.H.J.; Joosten, M.H.A.J. Soybean production in eastern and southern Africa and threat of yield loss due to soybean rust caused by Phakopsora pachyrhizi. Plant Pathol. 2016, 65, 176–188. [Google Scholar] [CrossRef]
- HEIL, M. Induced Systemic Resistance (ISR) Against Pathogens in the Context of Induced Plant Defences. Ann. Bot. 2002, 89, 503–512. [Google Scholar] [CrossRef] [Green Version]
- De Coninck, B.; Timmermans, P.; Vos, C.; Cammue, B.P.A.; Kazan, K. What lies beneath: Belowground defense strategies in plants. Trends Plant Sci. 2015, 20, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Labuschagne, N.; Dubery, I.A. The Chemistry of Plant–Microbe Interactions in the Rhizosphere and the Potential for Metabolomics to Reveal Signaling Related to Defense Priming and Induced Systemic Resistance. Front. Plant Sci. 2018, 9, 112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedras, M.S.C.; Abdoli, A. Pathogen inactivation of cruciferous phytoalexins: Detoxification reactions, enzymes and inhibitors. RSC Adv. 2017, 7, 23633–23646. [Google Scholar] [CrossRef] [Green Version]
- Mazid, M.; Khan, T.A.; Mohammad, F. Role of secondary metabolites in defense mechanisms of plants. Biol. Med. 2011, 3, 232–249. [Google Scholar]
- Castro-Moretti, F.R.; Gentzel, I.N.; Mackey, D.; Alonso, A.P. Metabolomics as an Emerging Tool for the Study of Plant–Pathogen Interactions. Metabolites 2020, 10, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunn, W.B.; Erban, A.; Weber, R.J.M.; Creek, D.J.; Brown, M.; Breitling, R.; Hankemeier, T.; Goodacre, R.; Neumann, S.; Kopka, J.; et al. Mass appeal: Metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics 2013, 9, 44–66. [Google Scholar] [CrossRef] [Green Version]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef]
- Glauser, G.; Veyrat, N.; Rochat, B.; Wolfender, J.L.; Turlings, T.C.J. Ultra-high pressure liquid chromatography-mass spectrometry for plant metabolomics: A systematic comparison of high-resolution quadrupole-time-of-flight and single stage Orbitrap mass spectrometers. J. Chromatogr. A 2013, 1292, 151–159. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metab. 2013, 1, 92–107. [Google Scholar] [CrossRef]
- Saccenti, E.; Hoefsloot, H.C.J.; Smilde, A.K.; Westerhuis, J.A.; Hendriks, M.M.W.B. Reflections on univariate and multivariate analysis of metabolomics data. Metabolomics 2014, 10, 361–374. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Silva, E.; da Graça, J.P.; Porto, C.; Martin do Prado, R.; Hoffmann-Campo, C.B.; Meyer, M.C.; de Oliveira Nunes, E.; Pilau, E.J. Unraveling Asian Soybean Rust metabolomics using mass spectrometry and Molecular Networking approach. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Miles, M.R.; Frederick, R.D.; Hartman, G.L. Evaluation of Soybean Germplasm for Resistance to Phakopsora pachyrhizi. Plant Health Prog. 2006, 7, 33. [Google Scholar] [CrossRef] [Green Version]
- Goellner, K.; Loehrer, M.; Langenbach, C.; Conrath, U.; Koch, E.; Schaffrath, U. Phakopsora pachyrhizi, the causal agent of Asian soybean rust. Mol. Plant Pathol. 2010, 11, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Gromski, P.S.; Muhamadali, H.; Ellis, D.I.; Xu, Y.; Correa, E.; Turner, M.L.; Goodacre, R. A tutorial review: Metabolomics and partial least squares-discriminant analysis–a marriage of convenience or a shotgun wedding. Anal. Chim. Acta 2015, 879, 10–23. [Google Scholar] [CrossRef]
- Triba, M.N.; Le Moyec, L.; Amathieu, R.; Goossens, C.; Bouchemal, N.; Nahon, P.; Rutledge, D.N.; Savarin, P. PLS/OPLS models in metabolomics: The impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol. Biosyst. 2015, 11, 13–19. [Google Scholar] [CrossRef]
- Ernst, M.; Kang, K.B.; Caraballo-Rodríguez, A.M.; Nothias, L.F.; Wandy, J.; Chen, C.; Wang, M.; Rogers, S.; Medema, M.H.; Dorrestein, P.C.; et al. Molnetenhancer: Enhanced molecular networks by integrating metabolome mining and annotation tools. Metabolites 2019, 9, 144. [Google Scholar] [CrossRef] [Green Version]
- da Silva, R.R.; Wang, M.; Nothias, L.F.; van der Hooft, J.J.J.; Caraballo-Rodríguez, A.M.; Fox, E.; Balunas, M.J.; Klassen, J.L.; Lopes, N.P.; Dorrestein, P.C. Propagating annotations of molecular networks using in silico fragmentation. PLoS Comput. Biol. 2018, 14, e1006089. [Google Scholar] [CrossRef]
- Wandy, J.; Zhu, Y.; Van Der Hooft, J.J.J.; Daly, R.; Barrett, M.P.; Rogers, S. Ms2lda.Org: Web-Based Topic Modelling for Substructure Discovery in Mass Spectrometry. Bioinformatics 2018, 34, 317–318. [Google Scholar] [CrossRef]
- Shah, J.; Chaturvedi, R. Lipid Signals in Plant-Pathogen Interactions. In Annual Plant Reviews Online; John Wiley & Sons, Ltd.: Chichester, UK, 2018; Volume 34, pp. 292–333. ISBN 9781119312994. [Google Scholar]
- Wasternack, C.; Feussner, I. The Oxylipin Pathways: Biochemistry and Function. Annu. Rev. Plant. Biol. 2018, 69, 363–386. [Google Scholar] [CrossRef]
- Ali, S. Jasmonic Acid Signaling Pathway in Response to Abiotic Stresses in Plants. Int. J. Mol. Sci. 2020, 21, 621. [Google Scholar] [CrossRef] [Green Version]
- Ishiga, Y.; Rao Uppalapati, S.; Gill, U.S.; Huhman, D.; Tang, Y.; Mysore, K.S. Transcriptomic and metabolomic analyses identify a role for chlorophyll catabolism and phytoalexin during Medicago nonhost resistance against Asian soybean rust. Sci. Rep. 2015, 5, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Loehrer, M.; Langenbach, C.; Goellner, K.; Conrath, U.; Schaffrath, U. Characterization of nonhost resistance of Arabidopsis to the asian soybean rust. Mol. Plant Microbe Interact. 2008, 21, 1421–1430. [Google Scholar] [CrossRef] [Green Version]
- Tzin, V.; Galili, G. The Biosynthetic Pathways for Shikimate and Aromatic Amino Acids in Arabidopsis thaliana. Arab. B 2010, 8. [Google Scholar] [CrossRef] [Green Version]
- Tohge, T.; Watanabe, M.; Hoefgen, R.; Fernie, A.R. Shikimate and phenylalanine biosynthesis in the green lineage. Front. Plant Sci. 2013, 4, 62. [Google Scholar] [CrossRef] [Green Version]
- Vogt, T. Phenylpropanoid Biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tremblay, A.; Hosseini, P.; Alkharouf, N.W.; Li, S.; Matthews, B.F. Transcriptome analysis of a compatible response by Glycine max to Phakopsora pachyrhizi infection. Plant Sci. 2010, 179, 183–193. [Google Scholar] [CrossRef]
- Morales, A.M.A.P.; O’rourke, J.A.; Van De Mortel, M.; Scheider, K.T.; Bancroft, T.J.; Borém, A.; Nelson, R.T.; Nettleton, D.; Baum, T.J.; Shoemaker, R.C.; et al. Transcriptome analyses and virus induced gene silencing identify genes in the Rpp4-mediated Asian soybean rust resistance pathway. Funct. Plant Biol. 2013, 40, 1029–1047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aoyagi, L.N.; Lopes-Caitar, V.S.; de Carvalho, M.C.C.G.; Darben, L.M.; Polizel-Podanosqui, A.; Kuwahara, M.K.; Nepomuceno, A.L.; Abdelnoor, R.V.; Marcelino-Guimarães, F.C. Genomic and transcriptomic characterization of the transcription factor family R2R3-MYB in soybean and its involvement in the resistance responses to Phakopsora pachyrhizi. Plant Sci. 2014, 229, 32–42. [Google Scholar] [CrossRef]
- Pandey, A.K.; Yang, C.; Zhang, C.; Graham, M.A.; Horstman, H.D.; Lee, Y.; Zabotina, O.A.; Hill, J.H.; Pedley, K.F.; Whitham, S.A. Functional analysis of the asian soybean rust resistance pathway mediated by Rpp2. Mol. Plant Microbe Interact. 2011, 24, 194–206. [Google Scholar] [CrossRef] [Green Version]
- Lygin, A.V.; Li, S.; Vittal, R.; Widholm, J.M.; Hartman, G.L.; Lozovaya, V.V. The importance of phenolic metabolism to limit the growth of Phakopsora pachyrhizi. Phytopathology 2009, 99, 1412–1420. [Google Scholar] [CrossRef] [Green Version]
- Stringlis, I.A.; De Jonge, R.; Pieterse, C.M.J. The Age of Coumarins in Plant-Microbe Interactions. Plant Cell Physiol. 2019, 60, 1405–1419. [Google Scholar] [CrossRef] [Green Version]
- De Menezes, H.D.; Pereira, A.C.; Brancini, G.T.P.; De Leão, H.C.; Massola Júnior, N.S.; Bachmann, L.; Wainwright, M.; Bastos, J.K.; Braga, G.U.L. Furocoumarins and coumarins photoinactivate Colletotrichum acutatum and Aspergillus nidulans fungi under solar radiation. J. Photochem. Photobiol. B Biol. 2014, 131, 74–83. [Google Scholar] [CrossRef]
- Flors, C.; Nonell, S. Light and Singlet Oxygen in Plant Defense Against Pathogens: Phototoxic Phenalenone Phytoalexins. ChemInform 2006, 39, 293–300. [Google Scholar] [CrossRef]
- Fracarolli, L.; Rodrigues, G.B.; Pereira, A.C.; Massola Júnior, N.S.; Silva-Junior, G.J.; Bachmann, L.; Wainwright, M.; Bastos, J.K.; Braga, G.U.L. Inactivation of plant-pathogenic fungus Colletotrichum acutatum with natural plant-produced photosensitizers under solar radiation. J. Photochem. Photobiol. B Biol. 2016, 162, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Beyer, S.F.; Beesley, A.; Rohmann, P.F.W.; Schultheiss, H.; Conrath, U.; Langenbach, C.J.G. The Arabidopsis non-host defence-associated coumarin scopoletin protects soybean from Asian soybean rust. Plant J. 2019, 99, 397–413. [Google Scholar] [CrossRef] [PubMed]
- Mert-Türk, F. Saponins versus plant fungal pathogens. J. Cell Mol. Biol. 2006, 5, 13–17. [Google Scholar]
- Papadopoulou, K.; Melton, R.E.; Leggett, M.; Daniels, M.J.; Osbourn, A.E. Compromised disease resistance in saponin-deficient plants. Proc. Natl. Acad. Sci. USA 1999, 96, 12923–12928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zehavi, U.; Ziv-Fecht, O.; Levy, M.; Naim, M.; Evron, R.; Polacheck, I. Synthesis and antifungal activity of medicagenic acid saponins on plant pathogens: Modification of the saccharide moiety and the 23α substitution. Carbohydr. Res. 1993, 244, 161–169. [Google Scholar] [CrossRef]
| Putative Metabolite Identification | Molecular Formula | [M + H]+ Measured | [M + H]+ Theoretical | Mass Accuracy (ppm) |
|---|---|---|---|---|
| Amino acid | ||||
| Proline | C5H9NO2 | 116.0704 | 116.0706 | −1.7 |
| Pipecolic acid | C6H11NO2 | 130.0861 | 130.0863 | −1.5 |
| Leucine | C6H13NO2 | 132.1020 | 132.1019 | 0.8 |
| Phenylalanine | C9H11NO2 | 166.0859 | 166.0863 | −2.4 |
| Tyrosine | C9H11NO3 | 182.0807 | 182.0812 | −2.7 |
| Tryptophan | C11H12N2O2 | 205.0964 | 205.0972 | −3.9 |
| Phenylpropanoids | ||||
| p-Coumaric acid | C9H8O3 | 165.0541 | 165.0546 | −3.0 |
| Citric acid | C6H8O7 | 193.0337 | 193.0343 | −3.1 |
| Ferulic acid | C10H10O4 | 195.0647 | 195.0642 | 2.6 |
| Abscisic acid | C15H20O4 | 265.1425 | 265.1434 | −3.4 |
| Peptides | ||||
| Ile-Pro | C11H20N2O3 | 229.1542 | 229.1546 | −1.7 |
| Ile-Val | C11H22N2O3 | 231.1700 | 231.1703 | −1.3 |
| Leu-Leu | C12H24N2O3 | 245.1855 | 245.1859 | −1.6 |
| Leu-Asn | C10H19N3O4 | 246.1439 | 246.1448 | −3.7 |
| Asp-Leu | C10H18N2O5 | 247.1279 | 247.1288 | −3.6 |
| Pro-Phe | C15H22N2O3 | 263.1412 | 263.1390 | 8.4 |
| Phe-Val | C14H20N2O3 | 265.1543 | 265.1546 | −1.1 |
| Leu-Phe | C14H18N2O3 | 279.1694 | 279.1703 | −3.2 |
| Asn-Phe | C13H17N3O4 | 280.1292 | 280.1291 | 0.4 |
| Leu-Leu-Gly | C14H27N3O4 | 302.2067 | 302.2074 | −2.3 |
| Peptide | C15H27N3O4 | 314.2066 | 314.2074 | −2.5 |
| Leu-Val-Val | C16H31N3O4 | 330.2377 | 330.2387 | −3.0 |
| Leu-Leu-Val | C17H33N3O4 | 344.2533 | 344.2543 | −2.9 |
| Cumarin | ||||
| 7-Methoxycoumarin | C10H8O3 | 177.0541 | 177.0546 | −2.8 |
| Scopoletin | C10H8O4 | 193.0488 | 193.0495 | −3.6 |
| Xanthyletin | C14H12O3 | 229.0857 | 229.0859 | −0.9 |
| Osthole | C15H16O3 | 245.1168 | 245.1172 | −1.6 |
| Flavonoids | ||||
| Daidzin | C21H20O9 | 417.1195 | 417.1180 | 3.6 |
| Daidzein | C15H10O4 | 255.0650 | 255.0651 | −0.4 |
| Neobavaisoflavone | C20H18O4 | 323.1273 | 323.1278 | −1.5 |
| Sojagol | C20H16O5 | 337.1067 | 337.1070 | −0.9 |
| Isoflavonoid | C21H20O4 | 337.1430 | 337.1434 | −1.2 |
| Gliceollin I | C20H18O5 | 339.1217 | 339.1227 | −2.9 |
| Gliceollin II | C20H18O5 | 339.1222 | 339.1227 | −1.5 |
| Gliceollin III | C20H18O5 | 339.1220 | 339.1227 | −2.1 |
| Isoflavonoid | C21H18O5 | 351.1219 | 351.1227 | −2.3 |
| Isoflavonoid | C21H20O5 | 353.1377 | 353.1383 | −1.7 |
| Xanthohumol | C21H22O5 | 355.1534 | 355.1540 | −1.7 |
| 7-O-Methylluteone | C21H20O6 | 369.1330 | 369.1333 | −0.7 |
| Schizandrin C | C22H24O6 | 385.1638 | 385.1645 | −1.8 |
| Genistin | C21H20O10 | 433.1118 | 433.1129 | −2.5 |
| Luteolin 8-C-glucoside | C21H21O11 | 449.1067 | 449.1078 | −2.4 |
| Isoquercitin | C21H20O12 | 465.1020 | 465.1027 | −1.5 |
| 3’-O-methyltricetin 3-O-α-L-rhamnopyranoside | C22H22O12 | 479.1170 | 479.1184 | −2.9 |
| Kaempferol-O-acetylhexoside | C23H22O12 | 491.1163 | 491.1184 | −4.3 |
| Malonyldaidzin | C24H22O12 | 503.1174 | 503.1184 | −2.0 |
| Formononetin 7-O-glucoside-6’’’’-O-malonate | C25H24O12 | 517.1339 | 517.1341 | −0.3 |
| Malonylgenistin | C24H22O13 | 519.1129 | 519.1133 | −0.8 |
| Isoorientin 2’’’’-O-rhamnoside | C27H30O15 | 595.1652 | 595.1657 | −0.8 |
| Rutin | C27H30O16 | 611.1588 | 611.1607 | −3.0 |
| Narcissin | C28H32O16 | 625.1747 | 625.1763 | −2.6 |
| Robinin | C33H40O19 | 741.2209 | 741.2236 | −3.6 |
| Flavonoid-7-O-glycosides | C33H40O20 | 757.2168 | 757.2186 | −2.3 |
| Flavonoid-7-O-glycosides | C34H42O20 | 771.2316 | 771.2342 | −3.4 |
| Lipids | ||||
| Jasmonic acid | C12H18O3 | 211.1331 | 211.1328 | 1.4 |
| (9Z,12Z,15Z)-octadeca-9,12,15-trien-6-ynoic acid | C18H26O2 | 275.2003 | 275.2005 | −0.7 |
| 13S-Hydroxy-9Z,11E,15Z-octadecatrienoic acid | C18H28O2 | 277.2153 | 277.2162 | −3.2 |
| Linolenic acid | C18H31O2 | 279.2318 | 279.2318 | 0.0 |
| 15-Methylhexadecasphinganine | C17H37NO2 | 288.2890 | 288.2897 | −2.4 |
| 12,13(S)-EOT | C18H28O3 | 293.2108 | 293.2111 | −1.0 |
| 12-OPDA | C18H28O3 | 293.2108 | 293.2111 | −1.0 |
| 10,13-Nonadecadiynoic acid | C19H30O2 | 291.2320 | 291.2318 | 0.7 |
| OPC-8:0 | C18H30O3 | 295.2266 | 295.2267 | −0.3 |
| 13-HPOT | C18H30O4 | 311.2213 | 311.2216 | −1.0 |
| Terpenes | ||||
| Soyasaponin III | C42H68O14 | 797.4641 | 797.4681 | −5.0 |
| Soyasaponin II | C47H76O17 | 913.5127 | 913.5155 | −3.1 |
| Dehydrosoyasaponin I | C48H76O18 | 941.5064 | 941.5104 | −4.2 |
| Soyasaponin I | C48H78O18 | 943.5243 | 943.5261 | −1.9 |
| Saponin | C55H70O14 | 955.4855 | 955.4838 | 1.8 |
| Asiaticoside | C48H78O19 | 959.5182 | 959.5210 | −2.9 |
| Cauloside D | C53H86O22 | 1075.5636 | 1075.5683 | −4.4 |
| Jujuboside B | C52H84O21 | 1045.5540 | 1045.5547 | −0.7 |
| Primulasaponin | C53H86O22 | 1105.5715 | 1105.5789 | −6.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, E.; Perez da Graça, J.; Porto, C.; Martin do Prado, R.; Nunes, E.; Corrêa Marcelino-Guimarães, F.; Conrado Meyer, M.; Jorge Pilau, E. Untargeted Metabolomics Analysis by UHPLC-MS/MS of Soybean Plant in a Compatible Response to Phakopsora pachyrhizi Infection. Metabolites 2021, 11, 179. https://doi.org/10.3390/metabo11030179
Silva E, Perez da Graça J, Porto C, Martin do Prado R, Nunes E, Corrêa Marcelino-Guimarães F, Conrado Meyer M, Jorge Pilau E. Untargeted Metabolomics Analysis by UHPLC-MS/MS of Soybean Plant in a Compatible Response to Phakopsora pachyrhizi Infection. Metabolites. 2021; 11(3):179. https://doi.org/10.3390/metabo11030179
Chicago/Turabian StyleSilva, Evandro, José Perez da Graça, Carla Porto, Rodolpho Martin do Prado, Estela Nunes, Francismar Corrêa Marcelino-Guimarães, Mauricio Conrado Meyer, and Eduardo Jorge Pilau. 2021. "Untargeted Metabolomics Analysis by UHPLC-MS/MS of Soybean Plant in a Compatible Response to Phakopsora pachyrhizi Infection" Metabolites 11, no. 3: 179. https://doi.org/10.3390/metabo11030179