Abstract
Aeromonas hydrophila RIT668 and Citrobacter freundii RIT669 were isolated from endangered spotted turtles (Clemmys guttata). Whole-genome sequencing, annotation and phylogenetic analyses of the genomes revealed that the closest relative of RIT668 is A. hydrophila ATCC 7966 and Citrobacter portucalensis A60 for RIT669. Resistome analysis showed that A. hydrophila and C. freundii harbor six and 19 different antibiotic resistance genes, respectively. Both bacteria colonize polyethylene and polypropylene, which are common plastics, found in the environment and are used to fabricate medical devices. The expression of six biofilm-related genes—biofilm peroxide resistance protein (bsmA), biofilm formation regulatory protein subunit R (bssR), biofilm formation regulatory protein subunit S (bssS), biofilm formation regulator (hmsP), toxin-antitoxin biofilm protein (tabA) and transcriptional activator of curli operon (csgD)—and two virulence factors—Vi antigen-related gene (viaB) and Shiga-like toxin (slt-II)—was investigated by RT-PCR. A. hydrophila displayed a > 2-fold increase in slt-II expression in cells adhering to both polymers, C. freundii adhering on polyethylene displayed a > 2-fold, and on polypropylene a > 6-fold upregulation of slt-II. Thus, the two new isolates are potential pathogens owing to their drug resistance, surface colonization and upregulation of a slt-II-type diarrheal toxin on polymer surfaces.
1. Introduction
Antibiotic resistance is an increasing crisis as both the range of resistance in clinical settings expands and the pipeline for development of new antibiotics contracts [1]. This problem is compounded by the global genomic scope of the antibiotic resistome, so that antibiotic resistance spans a continuum from genes in clinical pathogens to those of benign environmental microbes along with their proto-resistance gene progenitors [2,3]. Further, increased resistance to antimicrobial agents occurs in biofilms [4]. Biofilm-associated cells differ from their suspended counterparts by generation of an extracellular polymeric substance (EPS) matrix, reduced growth rates, and the up/downregulation of specific genes [5]. Plasmid conjugation occurs at a greater rate between cells in biofilms than between planktonic cells [6,7,8]. Bacterial biofilms constitute a serious problem for public health due to their potential to colonize in-dwelling medical devices (IMDs) [9,10], such as abdominal [11] and coronary stents [12], which contain polymeric materials such as polyethylene (PE) and polypropylene (PP). Once infected, the IMDs are often removed and replaced, causing a significant increase in the health care cost and chance of reinfection [13]. Similarly, water supplies contaminated with biofilms are a significant risk for public health [14].
Several studies reported the existence of this plastic-specific microbial community in aquatic systems [15,16,17,18,19,20,21]. For instance, the microbial community composition of the biofilm developing on submerged wood, glass and cellulose differed from that on PE and PP [16]. Characklis et al. [22], noted that the extent of microbial colonization appears to increase with surface roughness, and other reports show that microorganisms attach more rapidly to hydrophobic, non-polar surfaces such as Teflon and other plastics than to hydrophilic materials such as glass or metals [23,24,25]. The type of polymer influences the microbial community composition of the associated biofilm. For example, keratin-based biopolymers are being extensively explored for biomedical applications [26,27,28]. However, they may also be more susceptible to bacterial colonization, since Proteobacteria have been shown to colonize keratin-rich surfaces such as the shells of freshwater turtles [29]. Success in the war against biofilms requires a deeper understanding of the interactions between biofilm cells, the surface, antibiotics, and the host [30,31].
Aeromonas species are Gram-negative bacteria found in aquatic environments, soil, sewage and sediments. They are regarded as opportunistic pathogens of humans and other animals, especially fish. They can cause gastroenteritis, bacteraemia, peritonitis and infections of the respiratory system, hepato-biliary system, urinary tract, eyes and wounds [32,33]. Necrotizing fasciitis has also been reported with A. hydrophila as the causative agent [34,35]. Outbreaks of diarrhea have also been reported [36]. Most aeromonads have type IV pili, which facilitate horizontal gene transfer via conjugation of mobile genetic elements between different bacterial species [37]. Shiga-type toxins were reported from clinical and environmental Aeromonas isolates, with many having highly sequence similarity to homologs in virulent Escherichia coli strains [38,39]. Other studies show that up to 73% of the environmental strains may be naturally competent for taking up extracellular DNA [40]. Acquired resistance increases the multidrug-resistant character of both environmental and clinical strains of Aeromonas [41]. Extended-spectrum β-lactamase genes were transferred via plasmids from enterobacteria (possibly from the human gut) to Aeromonas [42]. Transposons, integrons and plasmids may also confer resistance to β-lactams, quinolones, macrolides, tetracyclines, sulfonamides, and chloramphenicol [32,43].
Recent studies have indicated that although Aeromonas spp. cause a range of pathologies, they are emerging as an enteric pathogen of public health concern [44]. Results from diarrheal outbreak studies further show that the infective dose of Aeromonas is very low [45]. A. hydrophila has also been found in food samples and has the ability to form mixed biofilms with other pathogens [46]. Aeromonads colonize the surfaces and insides of plants and animals [47], as well as abiotic surfaces like sediment, steel, glass, and polyvinyl chloride [48,49,50,51]. Notably, Aeromonas spp. attach to surfaces and eventually form biofilms even when freely able to grow in water, since this may enable long-term persistence in aquatic environments [52]. In natural systems, these persisting adherent cells were genetically distinct from the cells which were actually free-floating in the water column [53]. Biofilms, however, provide an ideal niche for the exchange of plasmids [6,7,8]. Studies have already noted the expression of similar Shiga-toxins in both environmental and clinical isolates of Aeromonas [39]. The relative abundance of Aeromonas spp. increased in riverine microplastics, suggesting that they could use plastics as vectors [20]. Recent studies have demonstrated that cells adhering to surfaces but not inside biofilms have resistance profiles similar to biofilm cells [54,55].
Citrobacter freundii is a member of the genus Citrobacter, belonging to the Gram-negative family Enterobacteriaceae [56,57,58]. It is found in soil, water, food and as a commensal in the gastrointestinal tract of humans and other animals [56,57,59]. However, it is increasingly a nosocomial and environmental pathogen, causing pneumonia, diarrhea, urinary tract and bloodstream infections [59,60,61,62,63,64,65,66]. Food poisoning cases caused by C. freundii isolates have been reported [59,62]. Cases of neonatal infections in preterm infants are also known [67]. Antibiotic-resistant C. freundii strains have been increasing around the world, and extended β-lactamase and plasmid-mediated quinolone resistance have been documented [66,68,69,70,71,72]. C. freundii strains have been isolated from mixed biofilms from water supply systems, growing along with strains such as Agrobacterium tumefaciens, A. hydrophila, Enterobacter soli and Stenotrophomonas maltophilia [73]. The presence of C. freundii worsens existing Pseudomonas aeruginosa infections in murine models and also likely in patients with co-infection [74]. Further, biofilm infections containing enteroaggregative E. coli and aggregative C. freundii cause diarrhea; in this case, the interaction is mediated by putative F pili [75]. Foodborne outbreaks, hospital and pediatric outbreaks, some involving hemolysis in addition to diarrhea, have also been reported [76,77,78]. Genomic analysis of a cytotoxic and aggregative strain of C. freundii revealed that it had acquired potential virulence factors including 7 genomic islands, 2 fimbriae islands and a type VI secretion system island [59]. Major virulence factors from human isolates were described as Shiga-like toxins and heat-stable toxins [77,79]. Later studies showed that an 18-amino acid peptide of a clinical C. freundii isolate was identical to that of an E. coli Shiga-toxin [79,80]. Other studies also reported Shiga-like toxins with 99.5–100% sequence similarity to E. coli toxins [81]. A cholera toxin B subunit homolog was also reported from a clinical strain [82].
Reptile-associated bacteria may be capable of infecting warm-blooded mammals, since there is a previous case of reptile and clinical strains, including Aeromonas spp., being identical [83]. Recent zoonotic outbreaks such as the SARS-CoV-2 pandemic [84] underscore the need to assess and identify potential future pathways for animal–human disease transmission. Both A. hydrophila and C. freundii are documented pathogens found among both freshwater and marine chelonians, and include antibiotic-resistant forms [85,86,87]. The study of antibiotic-resistant bacteria in marine turtles may also be used as a bio-indicator of exposure to effluents and other sources of environmental contamination [88,89]. Thus, turtles are a “sentinel species” for ecosystem health [90,91]. The often illegal and unregulated trade of reptiles sold at “wet markets” worldwide poses serious human health threats [92,93]. Physiologically stressed freshwater and marine reptiles are most often maintained in unsanitary conditions in plastic and wooden enclosures, providing ideal conditions for disease-causing bacteria and the formation of biofilms. Recent studies raised the possibility that immune-compromised turtles may be harboring and expressing significant pathogenic potential in the gut, and contribute to spreading them in the marine environment [90,94,95]. It is already known that marine turtles ingest microplastics [96] and that microplastics promote gene exchange and the occurrence of integrase genes such as int1 [97,98]. Connecting the dots, A. hydrophila and C. freundii could inhabit freshwater and marine reptiles and colonize the plastics they ingest. Then, highly drug-resistant biofilm forms of these bacteria could grow on the plastic surfaces within the gut and may be released by feces and contaminate water/food and eventually cause human disease. The possible colonization of plastic surfaces of water piping and IMD by turtle-derived A. hydrophila and C. freundii may carry additional risks of infection.
The aim of this study was to predict the potential for antibiotic resistance of our isolates based on whole-genome sequencing. Apart from antibiotic resistance genes, biofilm formation and the presence of toxins increase the damage any potential pathogen may cause. Therefore, we aimed to study the expression of six selected biofilm-related genes (bsmA, bssR, bssS, hmsP, tabA and csgD), and the virulence-associated viaB and slt-II during planktonic and adherent growth with PE and PP surfaces, after a period of 6 weeks. Genome analysis of our strains shows that both of them are potentially multidrug resistant. Further, we show that the expression of slt-II is enhanced in the adherent growth phase for both organisms on either PE or PP. We suggest that the predicted extensive antibiotic resistance and the ability to express a Shiga-like toxin warrants the classification of both strains as potential zoonotic opportunistic pathogens. Finally, the adherent phenotypes generated in this study were distinct compared to classical biofilms induced by the presence of blood/bile components, when analyzed by scanning electron microscopy.
2. Materials and Methods
2.1. Bacterial Isolation
Microbial samples were isolated from 12 adult rescued infected spotted turtles (Clemmys guttata) seized by the United States Fish and Wildlife Service from an illegal reptile trade operation (chain of custody ID number-ST#032797). The spotted turtle is a small, semi-aquatic, North American species commonly targeted and illegally harvested for sale in the pet trade and overseas for other uses [99,100]. The eyes, nostrils and limbs of turtles were swabbed on to agar plates and the samples were initially subjected to biochemical assays. For subsequent analysis, the two strains identified as Aeromonas hydrophila RIT 668 and Citrobacter freundii RIT 669 (based on 16S rDNA sequences) were routinely cultured on blood agar plates (5% sheep blood) and MacConkey plates respectively (BD BBL™, prepared media, 100 mm × 15 mm, San Diego, CA, USA). Hemolysis on the blood plates was examined by observing the presence of complete lysis around the colonies and a clearing on the medium. All twelve turtles (Clemmys guttata) were infected, lethargic and had reduced/stopped food intake for around a week. The eyes, nostrils and feet of the spotted turtles were surrounded by a slimy substance, and the eyes were inflamed. All the turtles were infected by A. hydrophila and C. freundii.
2.2. Characterization and Identification: Biochemical Assay and 16S rDNA Amplification
Primary identification was achieved via Gram staining. Oxidase and catalase tests were performed, followed by five groups of biochemical assays for microbial identification: Group 1—glucose, gas, and lysine; Group 2—ornithine, H2S, and indole; Group 3—adonitol, lactose, and arabinose; Group 4—sorbitol, Voges Proskauer, and dulcitol; and Group 5—phenylalanine (PA), urea, and citrate. Of the four different microbes identified, two strains were further analyzed and identified via 16S rDNA PCR amplification. For the 16S rDNA amplification, the microbial DNA was isolated using the ‘UltraClean Microbial DNA Isolation Kit’ (MO BIO Laboratories Inc., San Diego, CA, USA). Colony PCR was performed to obtain ~500 base pair amplicons of the 16S V3/V4 regions of the rRNA gene using the following conditions: 1 cycle at 95 °C for 2 min, followed by 30 cycles at 95 °C for 30 s, 52 °C for 30 s, and 72 °C for 3 min. The forward and reverse primers used for this was 5′-CCTACGGGNGGCWCGAG-3′ and 5′-GACTACHVGGGTATCTAATCC-3′. The amplicons were separated by gel electrophoresis, followed by commercial Sanger nucleotide sequencing (GeneWiz LLC, South Plainfield, NJ, USA) using the V3/V4 forward primer.
2.3. Genomic DNA Isolation
Genomic DNA was extracted from mid-log phase of Aeromonas hydrophila RIT668 and Citrobacter fruendii RIT669 using the GenElute Bacterial Genomic DNA kit, as per the manufacturer’s protocol (Sigma-Aldrich, St. Louis, MO, USA), and quantified using a Nanodrop One spectrophotometer.
2.4. Agarose Gel Electrophoreses
The DNA fragments from PCR and RT-PCR experiments were run on 1% agarose gels submerged in 1X TAE buffer, and stained with ethidium bromide, followed by visualization with a UV light source. A 1 kb ladder was used as the DNA standard (New England Biolabs, Ipswich, MA, USA).
2.5. Whole-Genome Sequencing, Assembly and Annotation
Using the Nextera XT library prep kit (Illumina) and Nextera XT index kit (Illumina), 1 ng of genomic DNA was processed to generate a sequencing-ready library, as per the manufacturer’s protocol. After library prep, 5 µL tagmented and indexed library DNA was quantified via the PicoGreen broad range assay on a Biotek Synergy H1 plate reader. To determine the fragment size distribution of DNA in the tagmented and indexed library, 1 µL of DNA was run using the DNA 1000 chip kit on an Agilent 2100 bioanalyzer. Based on the DNA concentration and size distribution, the library was manually normalized to a concentration of 4 nM in molecular-grade water. Of the resulting 4 nM library, 5 µL was then pooled with other tagmented, indexed, and normalized libraries. The pooled libraries were denatured and diluted to a loading concentration of 12 pM following the manufacturer’s protocol for manually normalized libraries. The pooled, denatured and diluted 12 pM library was sequenced using the MiSeq Reagent Kit V3 on the Illumina MiSeq for 2 × 151 cycles at the Rochester Institute of Technology Genomics Facility. Adapter trimming was performed automatically on the Illumina MiSeq during FASTQ generation. Trimmed reads were uploaded to the Galaxy web platform, and assembled de novo at the public server at usegalaxy.org [101], using Unicycler version 0.4.6.0 [102], with a minimum contig length of 200 bp.
2.6. Phylogenetic Analysis
The assembled FASTA contig files for A. hydrophila RIT668 and C. freundii RIT669 were uploaded to the Type Strain Genome Server (TYGS), found at https://tygs.dsmz.de. TYGS is a freely available tool for creating taxonomic assignments based on whole-genome sequence data [103]. The ten closest strains in the TYGS database to the query assembly were determined using Mash, a whole-genome clustering method [104]. Additionally, another ten closest relatives to the query assembly were selected by BLAST comparison of the 16S rDNA sequences extracted from the query using RNAmmer [105] against all 10,342 type strains in the TYGS database [106], selecting the top 50 hits, and calculating the Genome BLAST Distance Phylogeny distance (GBDP) with the query genome to determine the closest 10 type strains [107].
2.7. Resistance Gene Identifier (RGI)
The antibiotic resistance genes in both the model organisms were identified using the Resistance Gene Identifier (RGI) tool for understanding the mechanisms of antimicrobial resistance. The AMR gene family was identified along with the drug class, percentage identify of matching region and length of reference sequence. The genomes of A. hydrophila and C. freundii were analyzed using the bioinformatics platform RGI CARD. FASTA files were uploaded to RGI CARD software server RGI 5.1.0, CARD 3.0.4 [108] using the following parameters: Perfect, Strict and Loose hits, and complete genes only. The Perfect algorithm is most useful for clinical surveillance as it detects perfect matches to the curated reference sequences and mutations in the CARD. The Strict algorithm is aimed at unearthing previously unknown variants of known antibiotic resistance genes, including secondary screen for key mutations.
2.8. Predictions of Secondary Metabolite Production
The assembled genome sequence of A. hydrophila RIT668 and C. freundii RIT669 were analyzed using the Antibiotics and Secondary Metabolite Analysis Shell (antiSMASH5.0) webserver [109]. This tool identifies the biosynthetic loci covering the whole range of known secondary metabolite compound classes. It aligns the identified regions at the gene cluster levels to the closest match from a database, which contains all other known gene clusters and integrates the previously available secondary metabolite-related genes.
2.9. Colonization of Planktonic and Biofilm Forms on Plastics
2.9.1. Classical Biofilm Forms
The colonization of polyethylene (PE) and polypropylene (PP) plastics was studied using the two model organisms. High-density 1.6 mm PE and PP sheets were laser cut to 1 inch squares and sterilized with alcohol followed by UV radiation for 3 h. Sterilized squares were placed aseptically in blood agar plates and MacConkey plates, and inoculated with 5 mL each of overnight culture of A. hydrophila and C. freundii, respectively, for biofilm formation. The plates were incubated at 37 °C for 6 weeks with a steady supply of 3 mL of liquid bacterial cultures and analyzed for plastic biofilm colonization via scanning electron microscopy (SEM). This set up in solid agar media is referred to as ‘biofilms’ throughout the study and was used only for SEM comparisons.
2.9.2. Planktonic and Adherent Forms
A single plastic PE or PP square was placed in each well in a six-well dish with 5 mL of C. freundii or A. hydrophila culture with an OD600 of 0.5. Planktonic cells were aspirated out every other day in order to replace the well with fresh TSB depending on the dish. Dishes were incubated at 37 °C. Controls were initially placed alongside cultures in a six-well dish. To allow for maximum possible plastic colonization, except for aspiration of old TSB media and addition of new media, the six-well dish was left undisturbed for the duration of 6 weeks. This set up in liquid media is referred to as ‘planktonic’ throughout the study and the adherent cells as ‘adherent’.
2.9.3. RNA Isolation
RNA was isolated from the A. hydrophila and C. freundii samples using the Omega E.Z.N.A. bacterial RNA isolation kit as per the manufacturers’ protocol (Omega Bio-tek Inc., Norcross, GA, USA). The RNA integrity was analyzed NanoDrop followed by running the samples in a 1.5% agarose gel alongside a 1 Kb ladder (Invitrogen, Carlsbad, CA, USA). An array of nine different annealing temperature were tested on eight different primers in order to elucidate and optimize the temperatures for each of the samples.
2.10. Detection of Biofilm-Related Genes and Virulence Factors by RT-PCR
RT-PCR was performed based on the primer sequences and melting temperatures listed in Table 4. The genes encoding the following proteins were analyzed (a) Biofilm-related BsmA (biofilm peroxide resistance protein), BssR (biofilm formation regulatory protein), BssS (biofilm formation regulatory protein), HmsP (biofilm formation regulator), TabA (toxin-antitoxin biofilm protein) and CsgD (transcriptional activator curli operon). (b) Virulence-related: ViaB (primers VIAB-1 and VIAB-2, expected product size 516 bp; [110]), and Shiga-like toxin II (SLT-II) using the published primers GK1 and GK4, designed to amplify slt-II genes with an expected product size of 1260 bp [81,111].
A reverse transcriptase system (2 step) with cDNA synthesis (Promega Corp., Madison, WI, USA) was used for RT-PCR followed by the thermal cycle program. Since nine different annealing temperatures were tested in order to optimize the temperatures for each of the samples and each primer, the thermal cycle program varied for the primers and samples used in this study. A sample thermal cycle program involved initial denaturation at 95 °C for 3 min ×denaturation, annealing and extension at varying temperatures for 1 min (35 cycles) and 72 °C for 1 min respectively, followed by a final extension at 72 °C for 5 min and a final hold at 4 °C.
2.11. Estimation of Gene Expression
The change in gene expression was analyzed by running electrophoresis gels with sample concentrations normalized. Then, each set of bands corresponding to amplified fragments was analyzed for intensity and peak area with background subtraction, using the scientific image analysis open platform ImageJ [112]. The values were then plotted in Microsoft Excel as bar graphs showing fold change normalized to the 16S gene values, in the form of bar graphs. The 16S gene was used as the housekeeping standard during the RT-PCR and its values were used in this analysis.
2.12. Scanning Electron Microscopy Analysis
The biofilm-covered PE and PP squares were gently rinsed with phosphate-buffered saline (PBS) buffer at pH 7.4 and 2% glutaraldehyde to fix the cells and then incubated at 25 °C for 90 min. The samples were dehydrated with ethanol, incubated, and rinsed successively (50% for ten minutes, 70% for ten minutes, 80% for ten minutes, 95% twice for ten minutes, and 100% three times for fifteen minutes) inside a Petri dish. All the liquid was removed and the Petri dish containing the samples was covered in parafilm and stored at 25 °C until further analysis. Samples were covered with gold-palladium for two minutes with an SPI sputter coater to mitigate charging in the electron beam. The SEM was performed at 5 kV using a Mira3Tescan field emission SEM at the Rochester Institute of Technology (RIT) Nano-Imaging Lab.
3. Results
3.1. Biochemical Characterization and Taxonomy
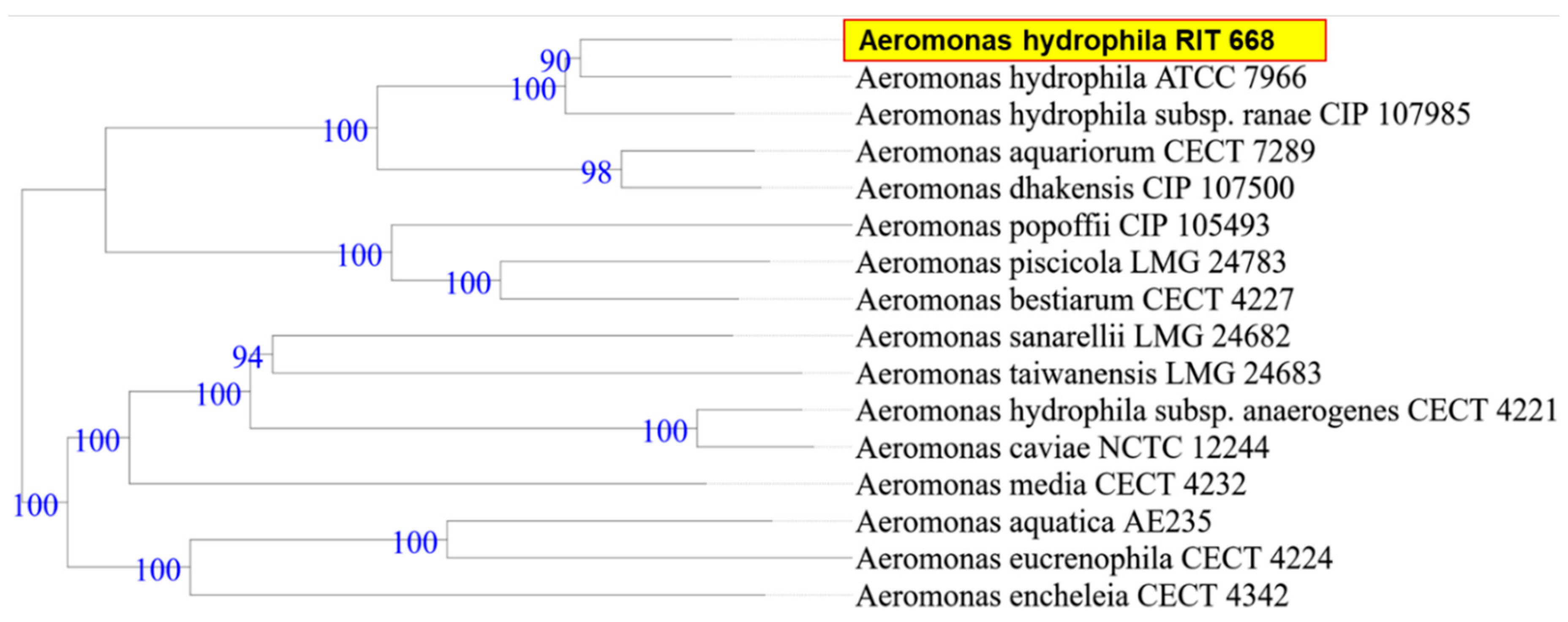
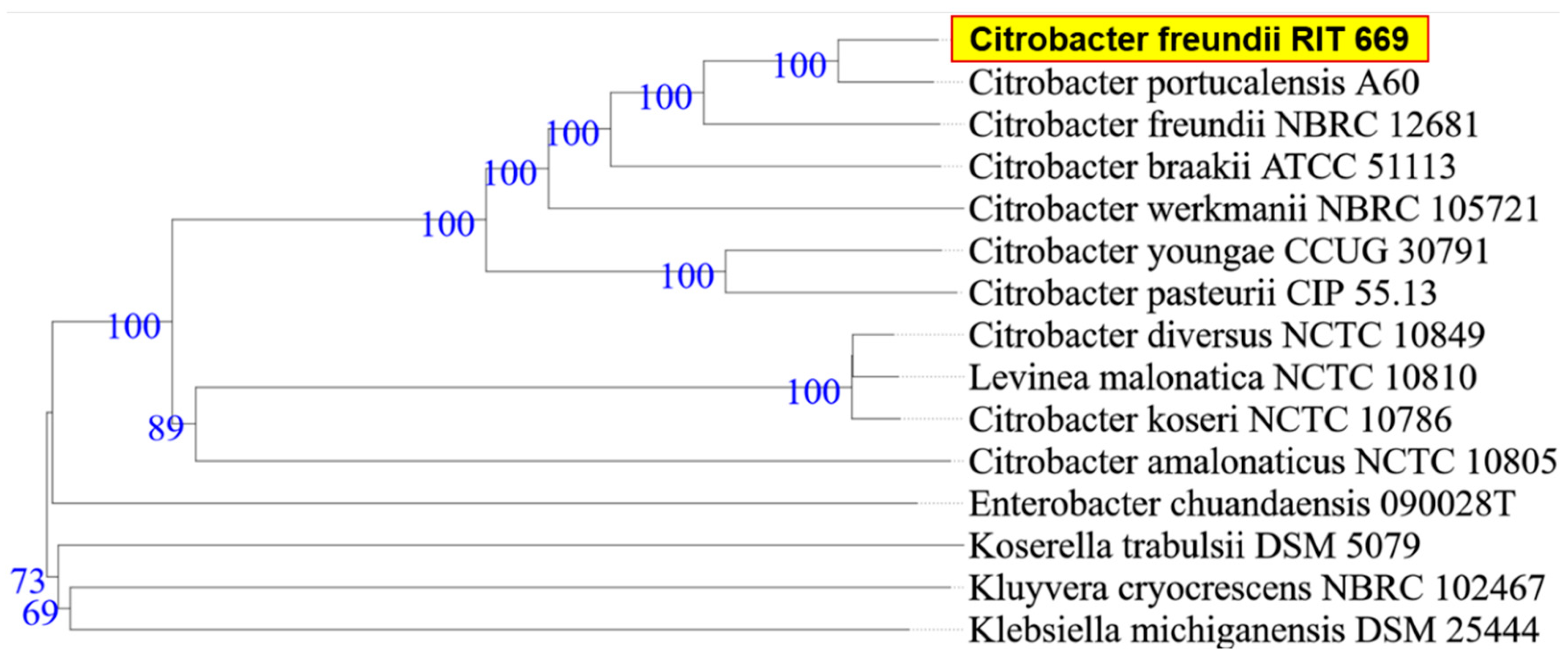
When the research was begun, there was 50% mortality, in spite of following appropriate animal care. By the end of the study period, however, all twelve turtles died (mortality rate 100%). Microbial isolates from rescued spotted turtles (Clemmys guttata) were identified as A. hydrophila and C. freundii, and were Gram-negative, beta hemolytic, lactose fermenting, and potential opportunistic pathogens. The two strains were initially identified through the 16S rDNA sequence coupled with NCBI-BLAST searches. Owing to their possible pathogenicity, the genomes were sequenced and annotated; a summary of the genome characteristics is shown in Table 1. Taxonomy based on whole-genome comparisons of the ten nearest relatives using the TYGS tool [103] confirmed the identification of RIT668 as A. hydrophila and RIT669 as Citrobacter. The nearest relative of A. hydrophila RIT668 is A. hydrophila ATCC 7966 (Figure 1). C. freundii RIT669 is the closest relative of Citrobacter portucalensis A60 (Figure 2).

Table 1.
Summary of whole-genome sequencing of A. hydrophila and C. freundii.
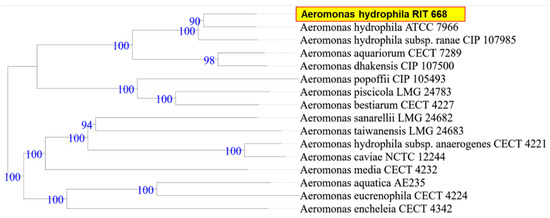
Figure 1.
WGS-based phylogenetic tree for A. hydrophila RIT668, using the TYGS platform. TYGS infers trees with FastME 2.1.4 [103] based on Genome BLAST Distance Phylogeny (GBDP) distances calculated from the genome sequences. Branch lengths are scaled in terms of GBDP distance formula d5 [107]. Numbers above branches are GBDP pseudo-bootstrap support values from 100 replications.
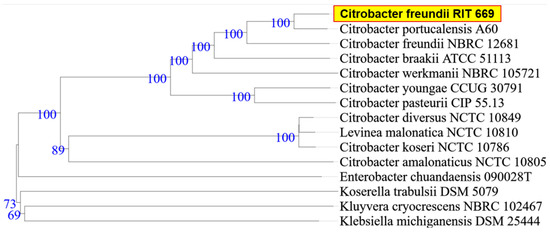
Figure 2.
WGS-based phylogenetic tree using TYGS for C. freundii RIT669. TYGS infers trees with FastME 2.1.4 [103] based on Genome BLAST Distance Phylogeny (GBDP) distances calculated from the genome sequences, whereas branch lengths are scaled in terms of GBDP distance formula d5 [107]. Numbers above branches are GBDP pseudo-bootstrap support values from 100 replications.
3.2. Resistome Analysis and Secondary Metabolite Analysis
According to the Resistance Gene Identifier (RGI) analysis of its genome, A. hydrophila RIT668 is potentially resistant to the carbapenem, penem, cephalosporin, fluroquinolone, tetracycline and elfamycin antibiotic classes (Table 2). It has homologs with 43–99.7% sequence identities to known examples of multidrug, unclassified, MLS (macrolide, lincosamide, streptogramin), aminoglycoside, β-lactam, bacitracin and glycopeptide resistance genes, with putative efflux pumps present. C. freundii RIT669 potentially resists up to 19 different classes of antibiotics, including carbapenem, penem, cephalosporin, fluroquinolone, tetracycline, peptides and elfamycin; mutilple efflux pumps are present (Table 3). Notably, it may also be resistant to classes such as fosfomycin, rhodamine, triclosan and benzalkonium chloride (the last two are widely used in consumer and sanitizer products). The closest relative C. portucalensis A60 is also aquatic and harbors resistance to β-lactams and quinolones [113].

Table 2.
Resistance Gene Identifier (RGI) analysis for A. hydrophila.

Table 3.
Resistance Gene Identifier (RGI) analysis for C. freundii.
3.3. Polymer Adhesion and Gene Expression
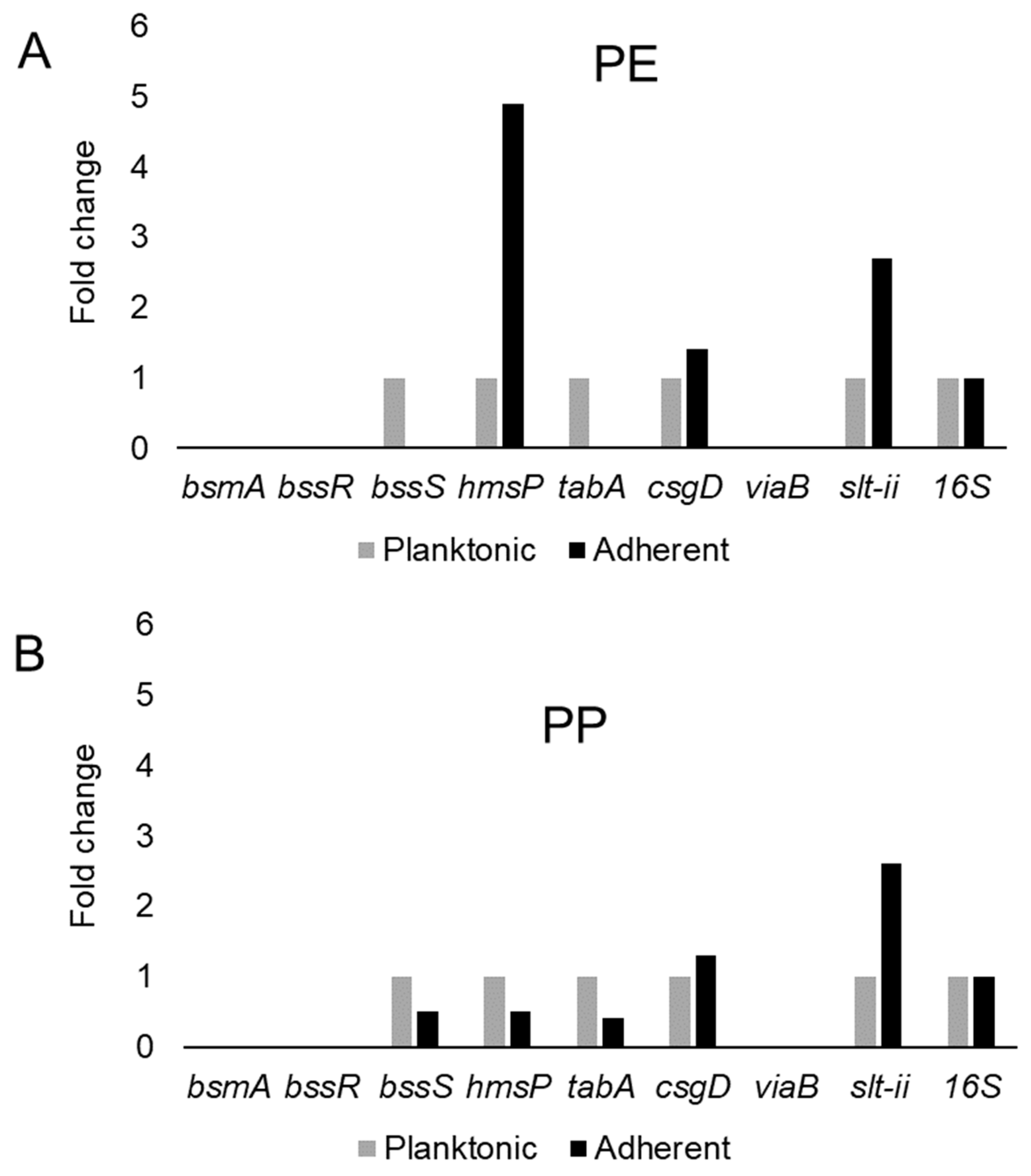
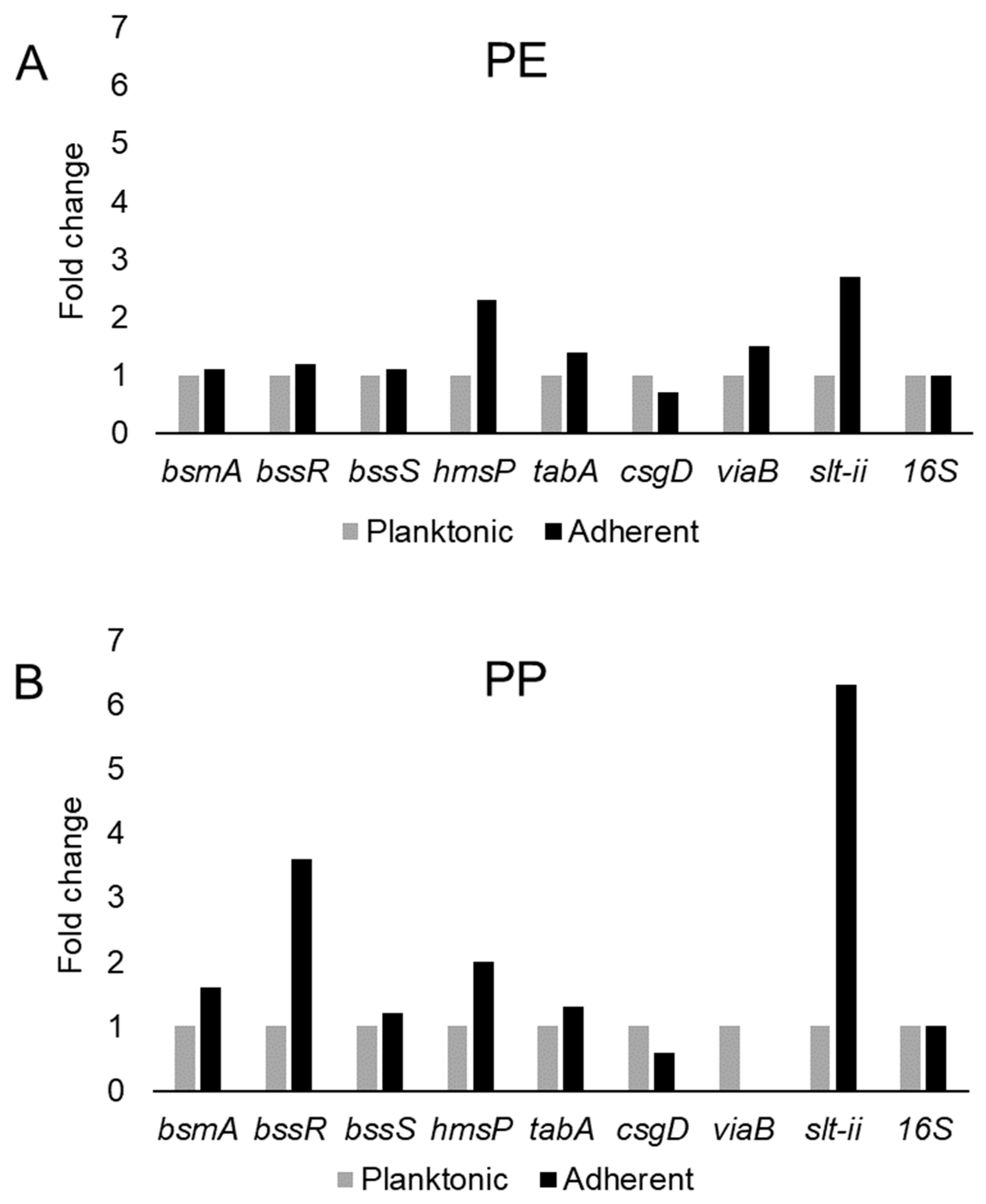
This study examined the expression of six biofilm-related genes as well as slt-II and viaB, based on the primers listed in Table 4, which were derived from earlier studies [81,85,114,115]. The genes bsmA, bssR, bssS and csgD were reported to contribute to planktonic and biofilm growth in Citrobacter werkmanii BF6 [114]. The genes tabA and hmsP were identified as biofilm related in a subsequent study on the same organism [115]. The gene bsmA (biofilm stress and motility) or biofilm peroxide resistance protein, known as yjfO in E. coli, is upregulated during biofilm growth [116] Mutant analysis revealed roles in microcolony formation, flagellar motility as well as resistance to acid and peroxide stresses [116]. It has been suggested that bsmA is involved in controlling cell aggregation at specific points in biofilm development [116,117]. The genes bssR and bssS (regulator of biofilm through signal secretion), also called yliH and yceP in E. coli K-12, were shown to repress motility by mediating cell signaling [118]. Both bssR and bssS were postulated to be global regulators of several genes involved in catabolite repression, stress response, quorum sensing and the putative stationary-phase signal [118]. A. hydrophila samples did not amplify PCR products corresponding to bsmA and bssR. However, planktonic cells showed the amplification of bssS, while cells adhering on PE did not express bssS and adherent cells on PP appeared to repress bssS expression (Figure 3). C. freundii expressed bsmA, bssR and bssS; most were slightly upregulated in adherent cells on both polymers, but bssR was upregulated over 3.5-fold in the PP-adherent cells (Figure 4).

Table 4.
Gene annotations and primer sequences of the biofilm-related and virulence genes in A. hydrophila and C. freundii.
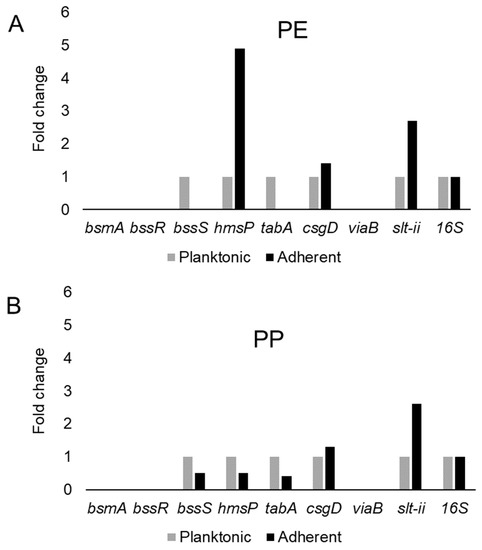
Figure 3.
Variation in gene expression during planktonic and adherent growth of A. hydrophila using tryptic soy broth on polyethylene (PE) surface as shown in (A) and polypropylene (PP) surface as shown in (B), respectively. The 16S rRNA gene expression was used as an internal control.
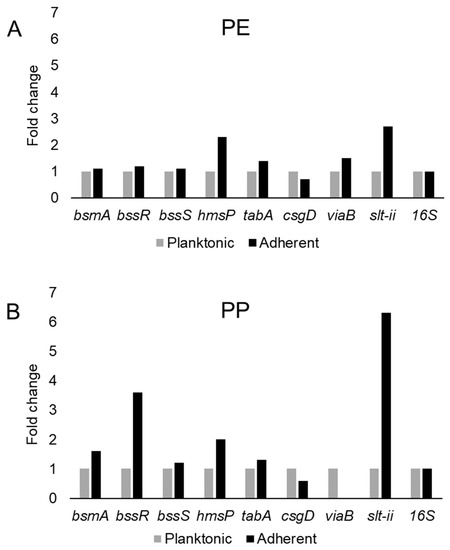
Figure 4.
Variation in gene expression during planktonic and adherent growth of C. freundii in tryptic soy broth on polyethylene (PE) surface as shown in (A) and polypropylene (PP) surface as shown in (B), respectively. The 16S rRNA gene expression was used as an internal control.
The gene hmsP (biofilm formation regulator) is one of two genes controlling the amount of biofilm produced in the pathogen Yersinia pestis via possible phosphodiesterase activity to modulate the levels of cyclic nucleotides [119]. It was shown that the EAL domain is essential for the biofilm inhibition activity of hmsP [120]. For A. hydrophila, hmsP was 4-fold upregulated in adherent cells on PE, but downregulated upon adherence to PP (Figure 3). C. freundii upregulated hmsP about 2-fold upon adherence to either PE or PP (Figure 4). The toxin-antitoxin gene tabA, known as yjgK in E. coli, influences biofilm formation in a time-dependent manner [121]. During the early phase tabA expression suppresses biofilm formation, while in later phases, it increases biofilm formation; it also suppresses biofilm dispersal [121]. PE-adherent A. hydrophila did not amplify tabA, but PP-adherent samples showed a slight downregulation (Figure 3). C. freundii showed a slight upregulation of tabA upon attachment to either polymer (Figure 4).
The curli operon-related gene csgD is a master regulator of biofilm formation and activates the synthesis of curli fimbriae and EPS in E. coli; csgD suppresses cell motility, triggering biofilm formation [122]. A homolog of csgD is expressed in the irreversible step of biofilm formation in Actinobacillus pleuropneumoniae [123]. Bacterial invasion of host cells may be controlled by c-di-GMP via csgD [124]. Cellulose biosynthesis is promoted by csgD, which is the transcriptional activator for curli production [125]. For both polymers, adherent A. hydrophila showed slightly upregulated expression of csgD (Figure 3), whereas adherent C. freundii downregulated csgD (Figure 4). The Vi antigen is commonly found in strains of Salmonella enterica serovars Typhi and Paratyphi, where it contributes to bacterial virulence and pathogenesis [126,127,128]. However, viaB-related genes for parts of the Vi antigen, which may contribute to invasiveness, have also been reported from Salmonella enterica serotype Dublin, E. coli and Citrobacter strains [129,130]. A. hydrophila samples did not amplify viaB (Figure 3), but slight upregulation is seen in PE-adhering C. freundii cells, whereas the PP-adhering cells did not amplify this gene (Figure 4). A. hydophila showed more than 2-fold upregulation of slt-II upon attachment to PE or PP (Figure 3). PE-adhering C. freundii upregulated slt-II expression more than 2-fold, while PP-adhering cells upregulated the expression of the same gene more than 6-fold (Figure 4). Biofilm-related genes were not upregulated and the lack of correlation between the expression of biofilm-related genes and the toxin genes remains unexplained. In Escherichia coli O104:H4 expressing the stx2 gene, the correlation of the expression of other genes such as pgaA and aggR with stx2 expression is strain dependent [131].
3.4. Secondary Metabolite Production via antiSMASH
Genome mining for secondary metabolite-producing gene clusters using the antiSMASH tool (108) showed that A. hydrophila has the potential to produce non-ribosomal peptides (NRP), arylpolyene, homeserine lactone and bacteriocin type of molecules (Table 5). C. freundii, on the other hand, possesses two biosynthetic gene clusters predicted to produce arylpolyene and NRP-type molecules (Table 5). The A. hydrophila gene clusters have high similarity to those of other aeromonads. Interestingly, homeserine lactone signals are known to coordinate quorum sensing and biofilm formation in many Gram-negative bacteria [132,133,134].

Table 5.
Antibiotics and Secondary Metabolites Analysis Shell (antiSMASH) prediction of biosynthetic gene clusters involved in the synthesis of antibiotics and secondary metabolites in A. hydrophila and C. freundii.
3.5. Electron Microscopy Analysis
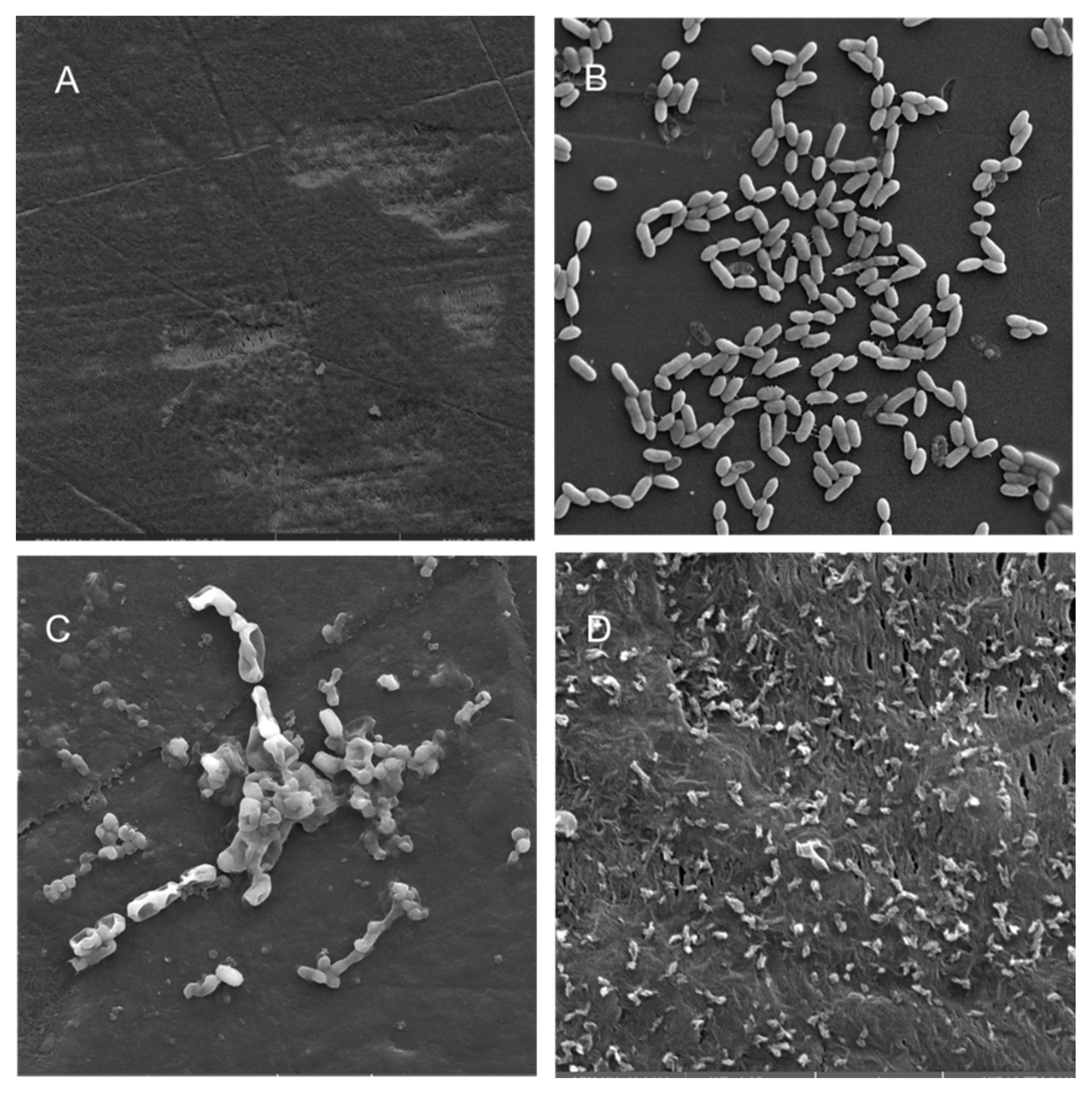
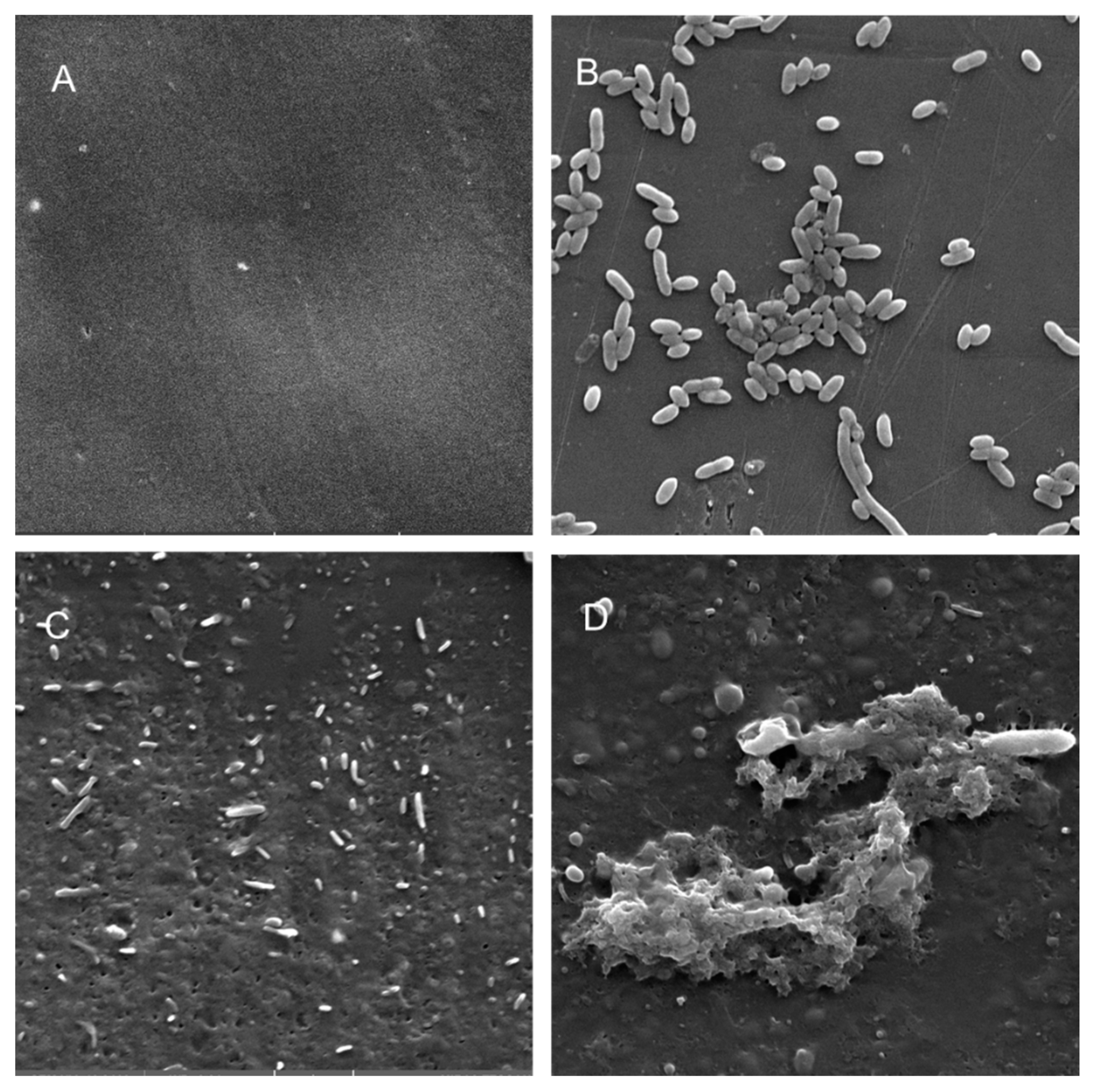
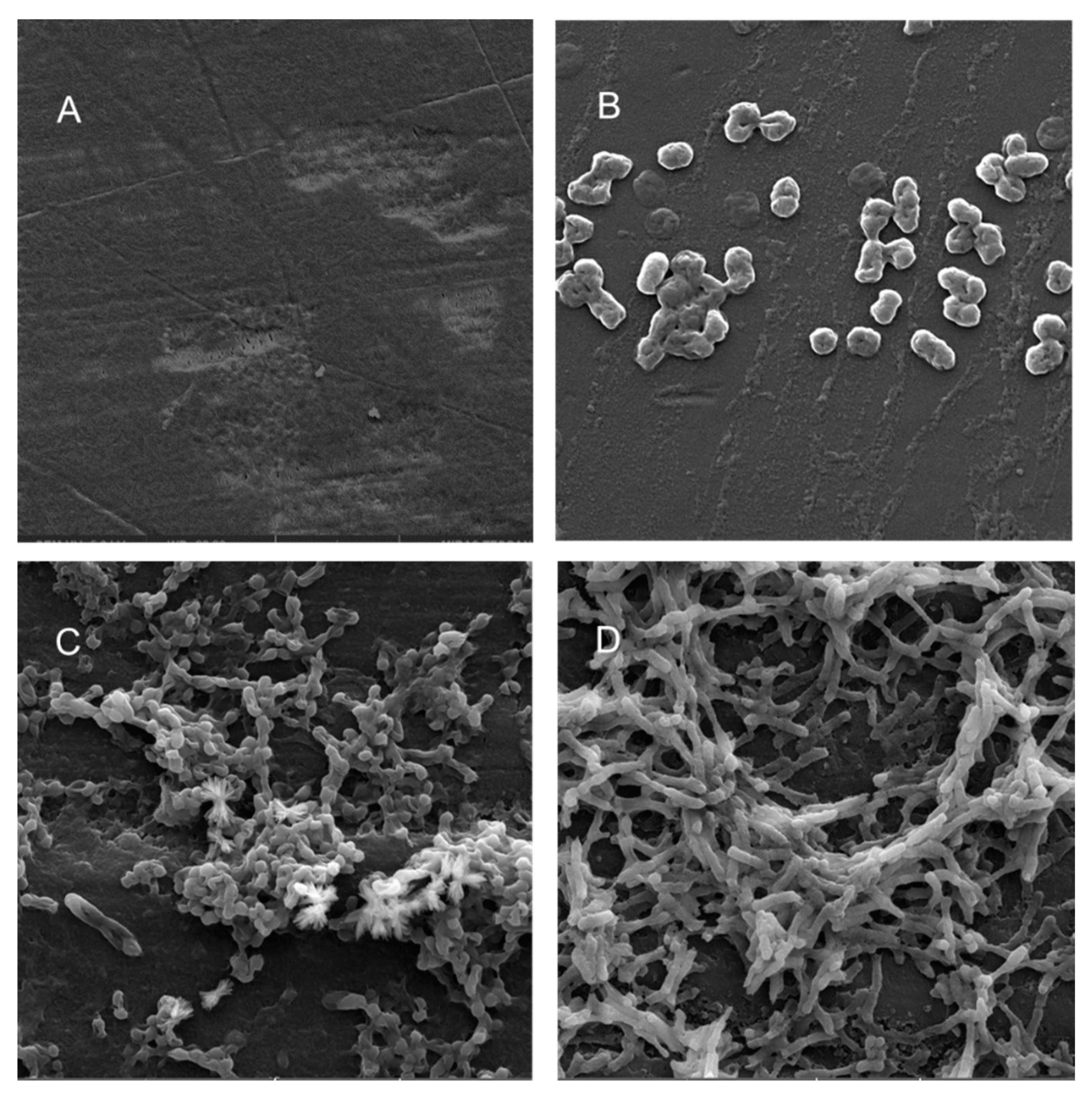
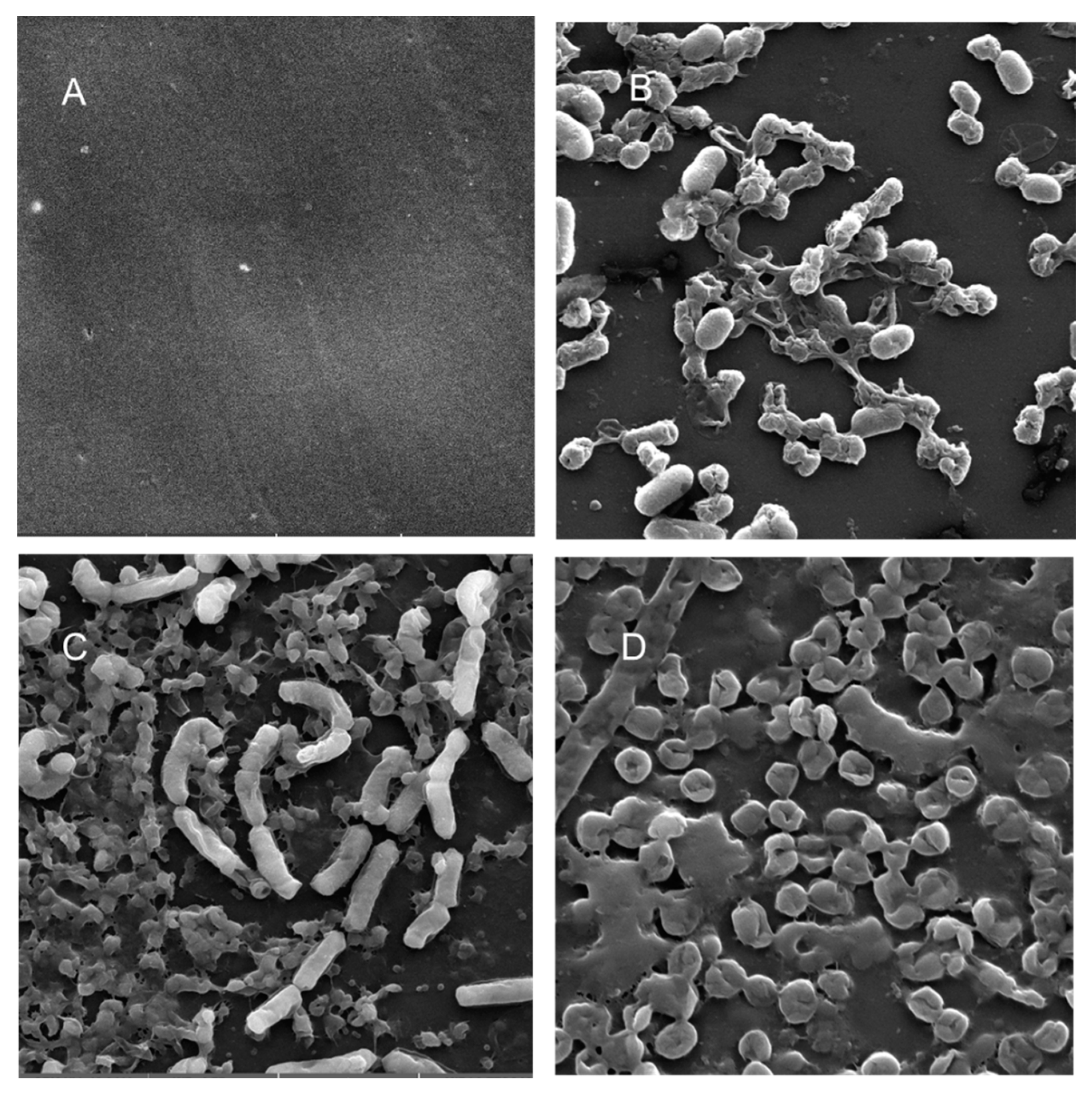
Scanning electron microscopy (SEM) was performed to compare the planktonic phases, adherent cells and blood/MacConkey media-induced classical biofilms. SEM analysis of A. hydrophila in the presence of PE showed that the long rod-shaped clumps of cells (Figure 5B) were transformed into long rows of cells adhered to the surface with some cell curling and the formation of early biofilm-like structures (Figure 5C). The mature biofilms induced by blood agar, on the other hand, had layers of EPS and only tiny remnants of cells visible (Figure 5D). For PP, the adherent cells had already formed EPS layers and only tiny cell fragments were visible (Figure 6C), but the blood agar induced the formation of irregularly shaped aerial structures (Figure 6D). For C. freundii, planktonic cells had a round morphology with clumping (Figure 7B), but the adherent cells already formed linked cells in a branching pattern and three-dimensional structures (Figure 7C). Mature biofilms induced by MacConkey medium appeared similar to the adherent cells, but were more densely networked (Figure 7D). C. freundii on adhered on PP differed significantly from earlier samples; here the cocci clumped together or linked by fibers (Figure 8B), are transformed into biofilms containing curled cells on top of cells trapped in EPS (Figure 8C). The MacConkey-induced mature biofilm was largely flat with cocci trapped in EPS on top (Figure 8D).
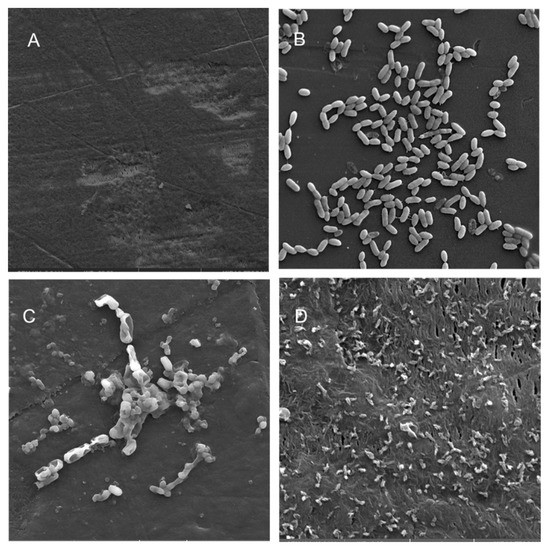
Figure 5.
Scanning electron micrographs (A) of un-colonized PE (×4980); (B–D) of A. hydrophila in the planktonic phase (×6470), adherent phase on PE (×15,900) and biofilm phase induced on PE pressed on blood agar (×19,700), respectively.
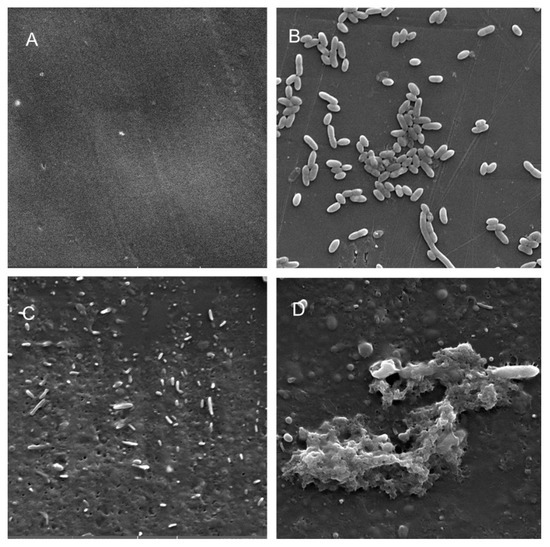
Figure 6.
Scanning electron micrographs (A) of un-colonized PP (×10,000); (B–D) of A. hydrophila in the planktonic phase (B; ×6540), adherent phase on PP (C; ×7530) and biofilm phase induced on PP pressed on blood agar (D; ×2540).
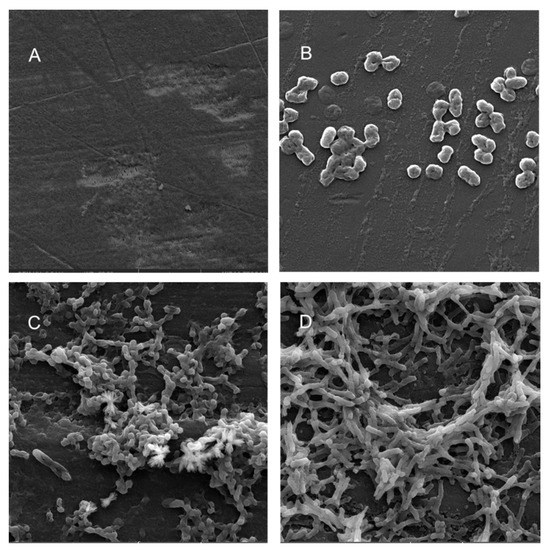
Figure 7.
Scanning electron micrographs (A) of un-colonized PE (×4980); (B–D) of C. freundii in the planktonic phase (×16,800), adherent phase on PE (×10,000) and biofilm phase induced on PE pressed on MacConkey agar (×8300), respectively.
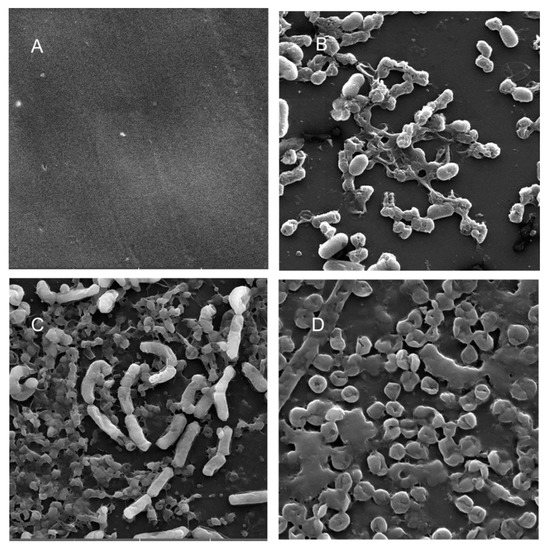
Figure 8.
Scanning electron micrographs (A) of un-colonized PP (×10,000); (B–D) of C. freundii in the planktonic phase (×18,700), adherent phase on PP (×11,200) and biofilm phase induced on PP pressed on MacConkey agar (×18,600), respectively.
4. Discussion
Genome analysis of the ATCC 7966-type strain (which is closely related to RIT668) showed it to be metabolically versatile with significant virulence potential and a predicted ability to infect a variety of hosts [134]. Due to the high similarity in the genomes (99%), it is possible that A. hydrophila RIT668 shares this potential for broad metabolic capability, virulence and the ability to infect multiple hosts, including humans. The closest relative of RIT669 is a C. portucalensis strain; C. portucalensis strains can be multidrug resistant and some may be highly resistant livestock-origin pathogens or “superbugs” [135,136]. This raises the possibility that RIT669 may also be multidrug resistant.
Enterobactericeae, especially Citrobacter spp., are common among turtles, whereby immune-compromised turtles may be conducive to the expression of pathogenic potential in the gut, which should be considered during rehabilitation procedures [94]. Australian green turtles (Chelonia mydas) in captivity were previously shown to harbor antibiotic-resistant strains [90,94,137]. A. hydrophila is common to infect turtles, fish and other amphibians as well as humans, as it is widely distributed in fresh water, estuarine and marine environments [138,139]. Among turtles, A. hydrophila infections were reported in Pseudemis scripta [140] and soft-shelled turtles (Trionyx sinensis) [141,142,143]. In the 1994 outbreak of A. hydrophila in Italy, there was a 95% mortality rate and the autopsies revealed infection in visceral organs [140]. The animals in that study were apathetic, did not feed and were lethargic in their movements. Our animals were similarly sluggish and the mortality rate was 100% by the end of the study.
Previously, it was shown that healthy pet turtles from seven species (other than Clemmys guttata) and their environment harbored Citrobacter spp. that were multidrug resistant, formed biofilms and were positive for slt-II and viaB [85]. In another study, all the turtles sampled had bacteria resistant to at least two antibiotics and 24% of the isolates were resistant to seven of the eight antibiotics tested [144]. Therefore, the predicted multidrug resistance characteristics of A. hydrophila RIT668 and C. freundii RIT669 are not surprising, and this fits into the growing number of studies which suggest that both A. hydrophila and C. freundii are emerging pathogens of concern. In addition, our strains do harbor more extensive resistomes in comparison to previous studies. Bacteria associated with reptiles could cross species barriers and infect mammals, according to a study of pet green turtles in which reptile and clinical strains for some enterobacterial genera (including Aeromonas) were identical [83]. We suggest that zoonotic transmission risks exist for these two potential pathogens and believe that their transmission needs to be monitored from a One Health perspective.
Shiga toxins (Stx) and Shiga-like toxins (Slt) are a group of bacterial toxins involved in human and other animal diseases; they are the cause of bloody diarrhea and hemolytic uremic syndrome [145,146]. Stx or Slt toxins are produced by enterohemorrhagic E. coli, Shigella dysenteriae type 1, C. freundii, Aeromonas spp. and Acinetobacter haemolyticus [38,39,81,145]. Diarrhea-associated C. freundii isolates are reported to contain several toxins, including Shiga-like toxins, heat-stable toxins and a homolog of the cholera toxin B subunit [59]. Clinical isolates of Aeromonas spp. possess Shiga toxin genes (stx1 and stx2), whose sequence is highly similar to the most virulent gene variants of E. coli strains [38]. It is notable that other studies showed specifically that microplastics in the environment may induce the overexpression of other types of virulence-related genes like the integrase int1 genes [98]. Similarly, the increased expression of the slt-II gene upon plastic colonization across all the samples suggests a possible adhesion-specific or even plastic adhesion-specific mechanism of upregulation. However, biofilm-related genes were not upregulated in this study and the lack of correlation between the expression of biofilm-related genes and the toxin genes remains unexplained. In E. coli O104:H4 expressing the stx2 gene, the correlation of the expression of other genes such as pgaA and aggR with stx2 expression is strain dependent [131]. Another study in the same E. coli strain, the only gene reported to be sufficient for plant surface adhesion is ompA, but the correlation is not strong [147]. Therefore, the co-regulated expression of biofilm and virulence-related genes is not consistent across different bacterial species, or even strains of the same species.
Electron microscopy has been used from the 1990s for the examination and characterization of biofilms on medical devices [148,149]. Scanning electron microscopy (SEM) has the level of magnification and resolution necessary to observe the overall shape of microorganisms in the biofilm, as well as their three-dimensional organization [150,151]. Aeromonas spp. were previously reported to attach to surfaces and eventually form biofilms, even if they were freely able to grow in water [52].
A recent study reported that floating biofilm-like structures (BLSs) and the attached biofilms had different metal resistance properties [152]. It has been suggested that “reversible” and “irreversible” attachment to a surface, as well as “surface-sentient” and “surface-naïve” planktonic cells are distinct [153]. The current work suggests that there could be different stages, and that adherent cells may have different properties from true biofilms. Thus, the adherent cells in a system with intermittent shear may not morphologically resemble the well-studied mature biofilm forms induced by blood/bile components, even though they may upregulate the expression of certain biofilm genes or toxins. Further, the structures formed in each case may differ based on both the bacterium and the polymer. Therefore, gene expression during different stages of attachment may be more nuanced than hitherto appreciated.
5. Conclusions
The isolated strains were shown to have significant resistomes, with A. hydrophila containing predicted resistance genes to six antibiotic classes and C. freundii containing resistance genes for 19 classes. The expression of many of the genes examined did not follow a specific pattern, since the adhesion to the polymers could be controlled via the complex interplay of several genes that were or were not included in this study. However, the clear exception is slt-II, whose expression is increased in response to either PE or PP for both bacteria. The toxin expression notwithstanding, electron microscopy showed that the adherent cells form structures different from well-studied biofilms growing on media with blood/bile components. Extensive antibiotic resistance repertoires, biofilm formation, colonization of common plastics and the overexpression of the slt-II-type diarrheal toxin in plastic-adherent cells, along with the origin in of the bacterial isolates from reptilian niches warrant the classification of both strains in this study as potential opportunistic zoonotic pathogens.
Author Contributions
The experimental work was performed by S.G.T. and M.A.G. The resistome and secondary metabolite analyses were performed by A.P. Whole-genome sequencing, annotation and phylogenetic analysis were performed by N.H.W., A.P. and S.G.T. planned the experiments. S.G.T., A.P., P.A.S. and A.O.H. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research and APC was funded by a National Institutes of Health (NIH) award (R15GM120653) to A.O.H.
Acknowledgments
The authors wish to thank Richard Hailstone for running the SEM samples.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Cooper, M.; Shlaes, D. Fix the antibiotics pipeline. Nature 2011, 472, 32. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. The antibiotic resistome. Expert Opin. Drug Discov. 2010, 5, 779–788. [Google Scholar] [CrossRef] [PubMed]
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic resistance is ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Ehlers, L.J.; Bouwer, E.J. RP4 Plasmid transfer among species of Pseudomonas in a biofilm reactor. Water Sci. Technol. 1999, 39, 163. [Google Scholar] [CrossRef]
- Roberts, A.P.; Pratten, J.; Wilson, M.; Mullany, P. Transfer of a Conjugative Transposon, Tn5397 in a Model Oral Biofilm. FEMS Microbiol. Lett. 1999, 177, 63–66. [Google Scholar] [CrossRef]
- Hausner, M.; Wuertz, S. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl. Environ. Microbiol. 1999, 65, 3710–3713. [Google Scholar] [CrossRef]
- Hancock, V.; Ferrières, L.; Klemm, P. Biofilm formation by asymptomatic and virulent urinary tract infectious Escherichia coli strains. FEMS Microbiol. Lett. 2007, 267, 30–37. [Google Scholar] [CrossRef]
- Shunmugaperumal, T. Biofilm Eradication and Prevention: A Pharmaceutical Approach to Medical Device Infections; John Wiley & Sons: Hoboken, NJ, USA, 2010; ISBN 978-0-470-47996-4. [Google Scholar]
- Jaff, M.R. Advances in the management of patients with vascular disease. Expert Rev. Cardiovasc. Ther. 2012, 10, 151–153. [Google Scholar] [CrossRef]
- Mani, G.; Feldman, M.; Patel, D.; Agrawal, C. Coronary stents: A materials perspective. Biomaterials 2002, 28, 1689–1710. [Google Scholar] [CrossRef] [PubMed]
- Engelsman, A.; Saldarriaga-Fernandez, I.; Nejadnik, M.; van Dam, G.; Francis, K.; Ploeg, R.; Busscher, H.; van der Mei, H. The risk of biomaterial-associated infection after revision surgery due to an experimental primary implant infection. Biofouling 2010, 26, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Ibekwe, A.M.; Murinda, S.E. Linking microbial community composition in treated wastewater with water quality in distribution systems and subsequent health effects. Microorganisms 2019, 7, 660. [Google Scholar] [CrossRef] [PubMed]
- De Tender, C.; Devriese, L.I.; Haegeman, A.; Maes, S.; Vangeyte, J.; Cattrijsse, A.; Dawyndt, P.; Ruttink, T. Temporal Dynamics of Bacterial and Fungal Colonization on Plastic Debris in the North Sea. Environ. Sci. Technol. 2017, 51, 7350–7360. [Google Scholar] [CrossRef] [PubMed]
- Kettner, M.T.; Rojas-Jimenez, K.; Oberbeckmann, S.; Labrenz, M.; Grossart, H.-P. Microplastics alter composition of fungal communities in aquatic ecosystems. Environ. Microbiol. 2017, 19, 4447–4459. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Wang, P.; Hou, J.; Yao, Y.; Liu, Z.; Liu, S.; Li, T. Distinct Community structure and microbial functions of biofilms colonizing microplastics. Sci. Total Environ. 2019, 650, 2395–2402. [Google Scholar] [CrossRef]
- Kirstein, I.V.; Wichels, A.; Krohne, G.; Gerdts, G. Mature biofilm communities on synthetic polymers in seawater-specific or general? Mar. Environ. Res. 2018, 142, 147–154. [Google Scholar] [CrossRef]
- Hoellein, T.J.; McCormick, A.R.; Hittie, J.; London, M.G.; Scott, J.W.; Kelly, J.J. Longitudinal patterns of microplastic concentration and bacterial assemblages in surface and benthic habitats of an urban river. Freshw. Sci. 2017, 36, 491–507. [Google Scholar] [CrossRef]
- McCormick, A.; Hoellein, T.J.; Mason, S.A.; Schluep, J.; Kelly, J.J. Microplastic is an abundant and distinct microbial habitat in an urban river. Environ. Sci. Technol. 2014, 48, 11863–11871. [Google Scholar] [CrossRef]
- Harrison, J.H.; Hoellein, T.J.; Sapp, M.; Tagg, A.S.; Ju-Nam, Y.; Ojeda, J.J. Microplastic-associated biofilms: A comparison of freshwater and marine environments. In Freshwater Microplastics; Wagner, M., Lambert, S., Eds.; Springer: Cham, Switzerland, 2018; Volume 58, pp. 181–201. [Google Scholar] [CrossRef]
- Characklis, W.; McFeters, G.; Marshall, K. Physiological ecology in biofilm systems. In Biofilms; Wiley Series in Ecological and Applied Microbiology; Characklis, W., Marshall, K., Eds.; John Wiley & Sons: New York, NY, USA, 1990; pp. 341–394. ISBN 9780471826637. [Google Scholar]
- Fletcher, M.; Loeb, G.I. Influence of substratum characteristics on the attachment of a marine pseudomonad to solid surfaces. Appl. Environ. Microbiol. 1979, 37, 67–72. [Google Scholar] [CrossRef]
- Pringle, J.H.; Fletcher, M. Influence of substratum wettability on attachment of freshwater bacteria to solid surfaces. Appl. Environ. Microbiol. 1983, 45, 811–817. [Google Scholar] [CrossRef] [PubMed]
- Bendinger, B.; Rijnaarts, H.H.; Altendorf, K.; Zehnder, A.J. Physicochemical cell surface and adhesive properties of coryneform bacteria related to the presence and chain length of mycolic acids. Appl. Environ. Microbiol. 1993, 59, 3973–3977. [Google Scholar] [CrossRef] [PubMed]
- Feroz, S.; Muhammad, N.; Ranayake, J.; Dias, G. Keratin-Based materials for biomedical applications. Bioact. Mater. 2020, 5, 496–509. [Google Scholar] [CrossRef] [PubMed]
- Mogosanu, G.D.; Grumezescu, A.M.; Chifiriuc, M.C. Keratin-Based biomaterials for biomedical applications. Curr. Drug Targets 2014, 15, 518–530. [Google Scholar] [CrossRef]
- Rajabi, M.; Ali, A.; McConnell, M.; Cabral, J. Keratinous materials: Structures and functions in biomedical applications. Mater. Sci. Eng. C 2020, 110, 110612. [Google Scholar] [CrossRef]
- McKnight, D.T.; Zenger, K.R.; Alford, R.A.; Huerlimann, R. Microbiome Diversity and Composition Varies Across Body Areas in a Freshwater Turtle. Microbiology 2020, 166, 440–452. [Google Scholar] [CrossRef]
- Costerton, J.; Stewart, P. Battling biofilms. Sci. Am. 2001, 285, 74–81. [Google Scholar] [CrossRef]
- Busscher, H.J.; van der Mei, H.C.; Subbiahdoss, G.; Jutte, P.C.; van den Dungen, J.J.; Zaat, S.A.; Schultz, M.J.; Grainger, D.W. Biomaterial-associated infection: Locating the finish line in the race for the surface. Sci. Transl. Med. 2012, 4, 153rv10. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. The genus aeromonas: Taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 2010, 23, 35–73. [Google Scholar] [CrossRef]
- Tena, D.; Aspiroz, C.; Figueras, M.J.; Gonzalez-Praetorius, A.; Aldea, M.J.; Alperi, A.; Bisquert, J. Surgical site infection due to Aeromonas species: Report of nine cases and literature review. Scand. J. Infect. Dis. 2009, 41, 164–170. [Google Scholar] [CrossRef]
- Minnaganti, V.R.; Patel, P.J.; Iancu, D.; Schoch, P.E.; Cunha, B.A. Necrotizing fasciitis caused by Aeromonas hydrophila. Heart Lung 2000, 29, 306–308. [Google Scholar] [CrossRef] [PubMed]
- Ugarte-Torres, A.; Perry, S.; Franko, A.; Church, D.L. Multidrug-resistant Aeromonas hydrophila causing fatal bilateral necrotizing fasciitis in an immunocompromised patient: A case report. J. Med. Case Rep. 2018, 12, 326. [Google Scholar] [CrossRef] [PubMed]
- Hochedez, P.; Hope-Rapp, E.; Olive, C.; Nicolas, M.; Beaucaire, G.; Cabié, A. Bacteremia caused by aeromonas species [corrected] complex in the Caribbean islands of Martinique and Guadeloupe. Am. J. Trop. Med. Hyg. 2010, 83, 1123–1127. [Google Scholar] [CrossRef] [PubMed]
- Rangrez, A.Y.; Dayananda, K.M.; Atanur, S.; Joshi, R.; Patole, M.S.; Shouche, Y.S. Detection of conjugation related type four secretion machinery in Aeromonas culicicola. PLoS ONE 2006, 1, e115. [Google Scholar] [CrossRef]
- Alperi, A.; Figueras, M.J. Human isolates of Aeromonas possess Shiga toxin genes (stx1 and stx2) highly similar to the most virulent gene variants of Escherichia coli. Clin. Microbiol. Infect. 2010, 16, 1563–1567. [Google Scholar] [CrossRef]
- Haque, Q.M.; Sugiyama, A.; Iwade, Y.; Midorikawa, Y.; Yamauchi, T. Diarrheal and environmental isolates of Aeromonas spp. Produce a toxin similar to Shiga-like toxin 1. Curr. Microbiol. 1996, 32, 239–245. [Google Scholar] [CrossRef]
- Huddleston, J.R.; Brokaw, J.M.; Zak, J.C.; Jeter, R.M. Natural transformation as a mechanism of horizontal gene transfer among environmental Aeromonas species. Syst. Appl. Microbiol. 2013, 36, 224–234. [Google Scholar] [CrossRef]
- Esteve, C.; Alcaide, E.; Giménez, M.J. Multidrug-resistant (MDR) Aeromonas Recovered from the metropolitan area of Valencia (Spain): Diseases spectrum and prevalence in the environment. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 137–145. [Google Scholar] [CrossRef]
- Marchandin, H.; Godreuil, S.; Darbas, H.; Jean-Pierre, H.; Jumas-Bilak, E.; Chanal, C.; Bonnet, R. Extended-spectrum beta-lactamase TEM-24 in an Aeromonas clinical strain: Acquisition from the prevalent Enterobacter aerogenes clone in France. Antimicrob. Agents Chemother. 2003, 47, 3994–3995. [Google Scholar] [CrossRef]
- Piotrowska, M.; Popowska, M. Insight into the mobilome of Aeromonas strains. Front. Microbiol. 2015, 6, 494. [Google Scholar] [CrossRef]
- Igbinosa, I.H.; Igumbor, E.U.; Aghdasi, F.; Tom, M.; Okoh, A.I. Emerging Aeromonas species infections and their significance in public health. Sci. World J. 2012, 2012, 625023. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Bravo, A.; Figueras, M.J. An update on the genus Aeromonas: Taxonomy, epidemiology, and pathogenicity. Microorganisms 2020, 8, 129. [Google Scholar] [CrossRef] [PubMed]
- Jahid, I.K.; Mizan, M.F.R.; Myoung, J.; Ha, S.-D. Aeromonas hydrophila biofilm, exoprotease, and quorum sensing responses to co-cultivation with diverse foodborne pathogens and food spoilage bacteria on crab surfaces. Biofouling 2018, 34, 1079–1092. [Google Scholar] [CrossRef] [PubMed]
- Mizan, M.F.; Jahid, I.K.; Ha, S.D. Microbial biofilms in seafood: A food-hygiene challenge. Food Microbiol. 2015, 49, 41–55. [Google Scholar] [CrossRef]
- Zalmum, A.A.; Marialegite, K.; Ghenghesh, K.S. Bacterial composition of the biofilm on the surface of course sediment of the danube: With special reference to the clinically important bacteria. Arch. Inst. Pasteur Tunis 1998, 75, 205–209. [Google Scholar]
- Balasubramanian, V.; Palanichamy, S.; Subramanian, G.; Rajaram, R. Development of polyvinyl chloride biofilms for succession of selected marine bacterial populations. J. Environ. Biol. 2012, 33, 57–60. [Google Scholar]
- Béchet, M.; Blondeau, R. Factors Associated with the Adherence and Biofilm Formation by Aeromonas caviae on Glass Surfaces. J. Appl. Microbiol. 2003, 94, 1072–1078. [Google Scholar] [CrossRef]
- Doğruöz, N.; Göksay, D.; Ilhan-Sungur, E.; Cotuk, A. Pioneer colonizer microorganisms in biofilm formation on galvanized steel in a simulated recirculating cooling-water system. J. Basic Microbiol. 2009, 49, S5–S12. [Google Scholar] [CrossRef]
- Kühn, I.; Allestam, G.; Huys, G.; Janssen, P.; Kersters, K.; Krovacek, K.; Stenström, T.A. Diversity, Persistence, and virulence of Aeromonas strains isolated from drinking water distribution systems in Sweden. Appl. Environ. Microbiol. 1997, 63, 2708–2715. [Google Scholar] [CrossRef]
- Villari, P.; Crispino, M.; Montuori, P.; Boccia, S. Molecular typing of Aeromonas isolates in natural mineral waters. Appl. Environ. Microbiol. 2003, 69, 697–701. [Google Scholar] [CrossRef]
- John, A.K.; Schmaler, M.; Khanna, N.; Landmann, R. Reversible daptomycin tolerance of adherent staphylococci in an implant infection model. Antimicrob. Agents Chemother. 2011, 55, 3510–3516. [Google Scholar] [CrossRef]
- Qu, Y.; Daley, A.J.; Istivan, T.S.; Rouch, D.A.; Deighton, M.A. Densely adherent growth mode, rather than extracellular polymer substance matrix build-up ability, contributes to high resistance of staphylococcus epidermidis biofilms to antibiotics. J. Antimicrob. Chemother. 2010, 65, 1405–1411. [Google Scholar] [CrossRef] [PubMed]
- Farmer, J. The genus citrobacter. In The Prokaryotes; Starr, M., Stolp, H., Truper, H., Balows, A., Schlegel, H., Eds.; Springer: Heidelberg, Germany, 1981; pp. 1140–1147. ISBN 978-3-662-13187-9. [Google Scholar]
- Sedlak, J. Citrobacter. In Enterobacteriaceae-Infektionen, Epidemiologie und Laboratoriumsdiagnostik; Sedlak, J., Rische, H., Eds.; VEB Georg Thieme Verlag: Leipzig, Germany, 1968; pp. 528–530. [Google Scholar]
- Sedlák, J. Present Knowledge and Aspects of Citrobacter. Curr Top. Microbiol. Immunol. 1973, 62, 41–59. [Google Scholar] [PubMed]
- Bai, L.; Xia, S.; Lan, R.; Liu, L.; Ye, C.; Wang, Y.; Jin, D.; Cui, Z.; Jing, H.; Xiong, Y.; et al. Isolation and characterization of cytotoxic, aggregative Citrobacter freundii. PLoS ONE 2012, 7, e33054. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.T.; Mitchell, L.A.; Zhao, L.; Mobley, H.L.T. Citrobacter freundii fitness during bloodstream Infection. Sci. Rep. 2018, 8, 11792. [Google Scholar] [CrossRef]
- Ranjan, K.P.; Ranjan, N. Citrobacter: An emerging health care associated urinary pathogen. Urol. Ann. 2013, 5, 313–314. [Google Scholar]
- Liu, L.; Chen, D.; Lan, R.; Hao, S.; Jin, W.; Sun, H.; Wang, Y.; Liang, Y.; Xu, J. Genetic diversity, multidrug resistance, and virulence of Citrobacter freundii from diarrheal patients and healthy individuals. Front. Cell. Infect. Microbiol. 2018, 8, 233. [Google Scholar] [CrossRef]
- Mohanty, S.; Singhal, R.; Sood, S.; Dhawan, B.; Kapil, A.; Das, B.K. Citrobacter infections in a tertiary care hospital in northern India. J. Infect. 2007, 54, 58–64. [Google Scholar] [CrossRef]
- Samonis, G.; Karageorgopoulos, D.E.; Kofteridis, D.P.; Matthaiou, D.K.; Sidiropoulou, V.; Maraki, S.; Falagas, M.E. Citrobacter infections in a general hospital: Characteristics and outcomes. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 61–68. [Google Scholar] [CrossRef]
- Chen, Y.S.; Wong, W.W.; Fung, C.P.; Yu, K.W.; Liu, C.Y. Clinical features and antimicrobial susceptibility trends in Citrobacter freundii bacteremia. J. Microbiol. Immunol. Infect. 2002, 35, 109–114. [Google Scholar]
- Liu, L.H.; Wang, N.Y.; Wu, A.Y.; Lin, C.C.; Lee, C.M.; Liu, C.P. Citrobacter freundii bacteremia: Risk factors of mortality and prevalence of resistance genes. J. Microbiol. Immunol. Infect. 2018, 51, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Brook, T.C.; Alcon-Giner, C.; Clarke, P.; Hall, L.J.; Hoyles, L. Draft genome sequences of Citrobacter freundii and Citrobacter murliniae strains isolated from the feces of preterm infants. Microbiol. Resour. Announc. 2019, 8, e00494-19. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Xiong, Z.; Li, X.; Hu, L.; Shen, J.; Li, T.; Hu, F.; Chen, S. Prevalence of plasmid-mediated quinolone resistance determinants in Citrobacter freundii isolates from Anhui Province, PR China. J. Med. Microbiol. 2011, 60, 1801–1805. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; Yu, J.K.; Lee, S.; Oh, E.J.; Woo, G.J. Prevalence and diversity of qnr alleles in AmpC-producing Enterobacter cloacae, Enterobacter aerogenes, Citrobacter freundii and Serratia marcescens: A multicentre study from Korea. J. Antimicrob. Chemother. 2007, 60, 868–871. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; Park, S.Y.; Oh, E.J.; Park, J.J.; Lee, K.Y.; Woo, G.J.; Lee, K. Occurrence of extended-spectrum beta-Lactamases among chromosomal AmpC-producing Enterobacter cloacae, Citrobacter freundii, and Serratia marcescens in Korea and investigation of screening criteria. Diagn. Microbiol. Infect. Dis. 2005, 51, 265–269. [Google Scholar] [CrossRef]
- Moland, E.S.; Hanson, N.D.; Black, J.A.; Hossain, A.; Song, W.; Thomson, K.S. Prevalence of newer beta-lactamases in gram-negative clinical isolates collected in the United States from 2001 to 2002. J. Clin. Microbiol. 2006, 44, 3318–3324. [Google Scholar] [CrossRef]
- Choi, S.H.; Lee, J.E.; Park, S.J.; Kim, M.N.; Choo, E.J.; Kwak, Y.G.; Jeong, J.Y.; Woo, J.H.; Kim, N.J.; Kim, Y.S. Prevalence, microbiology, and clinical characteristics of extended-spectrum beta-lactamase-producing Enterobacter spp., Serratia marcescens, Citrobacter freundii, and Morganella morganii in Korea. Eur. J. Clin. Microbiol. Infect. Dis. 2007, 26, 557–561. [Google Scholar] [CrossRef]
- Kregiel, D.; Rygala, A.; Kolesinska, B.; Nowacka, M.; Herc, A.S.; Kowalewska, A. Antimicrobial and antibiofilm N-acetyl-L-cysteine Grafted siloxane polymers with potential for use in water systems. Int. J. Mol. Sci. 2019, 20, 2011. [Google Scholar] [CrossRef]
- Peng, Q.; Chen, L.; Zhou, S.; Li, H.; Long, J.; Yao, F.; Zhuang, Y.; Zhang, Z.; Huang, Y.; Duan, K. Co-existence of Citrobacter freundii exacerbated Pseudomonas aeruginosa infection in vivo. Int. J. Med. Microbiol. 2020, 310, 151379. [Google Scholar] [CrossRef]
- Pereira, A.L.; Silva, T.N.; Gomes, A.C.; Araújo, A.C.; Giugliano, L.G. Diarrhea-associated biofilm formed by Enteroaggregative Escherichia coli and aggregative Citrobacter freundii: A Consortium mediated by putative F pili. BMC Microbiol. 2010, 10, 57. [Google Scholar] [CrossRef]
- Aminharati, F.; Ehrampoush, M.H.; Soltan Dallal, M.M.; Yaseri, M.; Dehghani Tafti, A.A.; Rajabi, Z. Citrobacter freundii foodborne disease outbreaks related to environmental conditions in Yazd Province, Iran. Iran. J. Public Health 2019, 48, 1099–1105. [Google Scholar] [CrossRef] [PubMed]
- Tschape, H.; Prager, R.; Streckel, W.; Fruth, A.; Tietze, E.; Böhme, G. Verotoxinogenic Citrobacter freundii associated with severe gastroenteritis and cases of haemolytic uraemic syndrome in a nursery school: Green butter as the infection source. Epidemiol. Infect. 1995, 114, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Parida, S.N.; Verma, I.C.; Deb, M.; Bhujwala, R.A. An outbreak of diarrhea due to Citrobacter freundii in a neonatal special care nursery. Indian J. Pediatr. 1980, 47, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Guarino, A.; Capano, G.; Malamisura, B.; Alessio, M.; Guandalini, S.; Rubino, A. Production of Escherichia coli STa-like heat-stable enterotoxin by Citrobacter freundii isolated from humans. J. Clin. Microbiol. 1987, 25, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Guarino, A.; Giannella, R.; Thompson, M.R. Citrobacter freundii produces an 18-Amino-acid heat-stable enterotoxin identical to the 18-amino-acid Escherichia coli heat-stable enterotoxin (ST Ia). Infect. Immun. 1989, 57, 649–652. [Google Scholar] [CrossRef]
- Schmidt, H.; Montag, M.; Bockemühl, J.; Heesemann, J.; Karch, H. Shiga-like toxin II-related cytotoxins in Citrobacter freundii strains from humans and beef samples. Infect. Immun. 1993, 61, 534–543. [Google Scholar] [CrossRef]
- Karasawa, T.; Ito, H.; Tsukamoto, T.; Yamasaki, S.; Kurazono, H.; Faruque, S.M.; Nair, G.B.; Nishibuchi, M.; Takeda, Y. Cloning and Characterization of genes encoding homologues of the B subunit of cholera toxin and the Escherichia coli heat-labile enterotoxin from clinical isolates of Citrobacter freundii and E. coli. Infect. Immun. 2002, 70, 7153–7155. [Google Scholar] [CrossRef]
- McCoy, R.H.; Seidler, R.J. Potential pathogens in the environment: Isolation, enumeration, and identification of seven genera of intestinal bacteria associated with small green pet turtles. Appl. Microbiol. 1973, 25, 534–538. [Google Scholar] [CrossRef]
- Murdoch, D.R.; French, N.P. COVID-19: Another infectious disease emerging at the animal-human interface. N. Z. Med. J. 2020, 133, 12–15. [Google Scholar]
- Hossain, S.; Wimalasena, S.H.M.P.; Heo, G.J. Virulence factors and antimicrobial resistance pattern of Citrobacter Freundii isolated from healthy pet turtles and their environment. Asian J. Anim. Vet. Adv. 2016, 12, 10–16. [Google Scholar] [CrossRef]
- Chung, T.; Yi, S.; Kim, B.; Kim, W.; Shin, G. Identification and antibiotic resistance profiling of bacterial isolates from septicaemic soft-shelled turtles (Pelodiscus sinensis). Vet. Med. 2017, 62, 169–177. [Google Scholar] [CrossRef]
- Wimalasena, S.H.M.P.; Shin, G.W.; Hossain, S.; Heo, G.J. Potential enterotoxicity and antimicrobial resistance pattern of Aeromonas species isolated from pet turtles and their environment. J. Vet. Med. Sci. 2017, 79, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Blasi, M.F.; Migliore, L.; Mattei, D.; Rotini, A.; Thaller, M.C.; Alduina, R. Antibiotic resistance of gram-negative bacteria from wild captured loggerhead sea turtles. Antibiotics 2020, 9, 162. [Google Scholar] [CrossRef]
- Al-Bahry, S.; Mahmoud, I.; Al-Zadjali, M.; Elshafie, A.; Al-Harthy, A.; Al-Alawi, W. Antibiotic resistant bacteria as bio-indicator of polluted effluent in the green turtles, Chelonia mydas in Oman. Mar. Environ. Res. 2011, 71, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Pace, A.; Dipineto, L.; Fioretti, A.; Hochscheid, S. Loggerhead sea turtles as sentinels in the western mediterranean: Antibiotic resistance and environment-related modifications of gram-negative bacteria. Mar. Pollut. Bull. 2019, 149, 110575. [Google Scholar] [CrossRef]
- Al-Bahry, S.N.; Al-Zadjali, M.A.; Mahmoud, I.Y.; Elshafie, A.E. Biomonitoring marine habitats in reference to antibiotic resistant bacteria and ampicillin resistance determinants from oviductal fluid of the nesting green sea turtle, Chelonia mydas. Chemosphere 2012, 87, 1308–1315. [Google Scholar] [CrossRef]
- Rabinowitz, P.; Conti, L. Links among human health, animal health, and ecosystem health. Annu. Rev. Public Health 2013, 34, 189–204. [Google Scholar] [CrossRef]
- Rosen, G.E.; Smith, K.F. Summarizing the evidence on the international trade in illegal wildlife. Ecohealth 2010, 7, 24–32. [Google Scholar] [CrossRef]
- Pace, A.; Rinaldi, L.; Ianniello, D.; Borrelli, L.; Cringoli, G.; Fioretti, A.; Hochscheid, S.; Dipineto, L. Gastrointestinal investigation of parasites and enterobacteriaceae in loggerhead sea turtles from italian coasts. BMC Vet. Res. 2019, 15, 370. [Google Scholar] [CrossRef]
- Alduina, R.; Gambino, D.; Presentato, A.; Gentile, A.; Sucato, A.; Savoca, D.; Filippello, S.; Visconti, G.; Caracappa, G.; Vicari, D.; et al. Is Caretta caretta a carrier of antibiotic resistance in the Mediterranean Sea? Antibiotics 2020, 9, 116. [Google Scholar] [CrossRef]
- Duncan, E.M.; Broderick, A.C.; Fuller, W.J.; Galloway, T.S.; Godfrey, M.H.; Hamann, M.; Limpus, C.J.; Lindeque, P.K.; Mayes, A.G.; Omeyer, L.C.M.; et al. Microplastic ingestion ubiquitous in marine turtles. Glob. Chang. Biol. 2019, 25, 744–752. [Google Scholar] [CrossRef] [PubMed]
- Arias-Andres, M.; Klümper, U.; Rojas-Jimenez, K.; Grossart, H.P. Microplastic pollution increases gene exchange in aquatic ecosystems. Environ. Pollut. 2018, 237, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Eckert, E.M.; Di Cesare, A.; Kettner, M.T.; Arias-Andres, M.; Fontaneto, D.; Grossart, H.P.; Corno, G. Microplastics increase impact of treated wastewater on freshwater microbial community. Environ. Pollut. 2018, 234, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Enneson, J.; Litzgus, J. Using long-term data and stage-classified matrix to assess conservation strategies for an endangered turtle (Clemmys guttata). Biol. Conserv. 2018, 141, 1560–1568. [Google Scholar] [CrossRef]
- Howell, J.; McKnight, D.; Seigel, R. A novel method of collecting spotted turtles (Clemmys guttata). Herpetol. Rev. 2016, 47, 28–31. [Google Scholar]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Lin, C.L.; Chiu, C.H.; Chu, C.; Huang, Y.C.; Lin, T.Y.; Ou, J.T. A Multiplex polymerase chain reaction method for rapid identification of Citrobacter freundii and Salmonella species, including Salmonella Typhi. J. Microbiol. Immunol. Infect. 2007, 40, 222–226. [Google Scholar]
- Schmitt, C.K.; McKee, M.L.; O’Brien, A.D. Two copies of shiga-like toxin ii-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157: H- strain E32511. Infect. Immun. 1991, 59, 1065–1073. [Google Scholar] [CrossRef]
- Schindelin, J.; Rueden, C.T.; Hiner, M.C.; Eliceiri, K.W. The imagej ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 2015, 82, 518–529. [Google Scholar] [CrossRef]
- Ribeiro, T.G.; Gonçalves, B.R.; da Silva, M.S.; Novais, Â.; Machado, E.; Carriço, J.A.; Peixe, L. Citrobacter portucalensis Sp. Nov., isolated from an aquatic sample. Int. J. Syst. Evol. Microbiol. 2017, 67, 3513–3517. [Google Scholar] [CrossRef]
- Zhou, G.; Li, L.J.; Shi, Q.S.; Ouyang, Y.S.; Chen, Y.B.; Hu, W.F. Effects of nutritional and environmental conditions on planktonic growth and biofilm formation of Citrobacter werkmanii BF-6. J. Microbiol. Biotechnol. 2013, 23, 1673–1682. [Google Scholar] [CrossRef]
- Zhou, G.; Peng, H.; Wang, Y.-S.; Huang, X.-M.; Xie, X.-B.; Shi, Q.-S. Complete genome sequence of Citrobacter werkmanii strain BF-6 isolated from industrial putrefaction. BMC Genom. 2017, 18, 765. [Google Scholar] [CrossRef]
- Weber, M.M.; French, C.L.; Barnes, M.B.; Siegele, D.A.; McLean, R.J. A previously uncharacterized gene, YjfO (BsmA), influences Escherichia coli biofilm formation and stress response. Microbiology 2010, 156, 139–147. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Labbate, M.; Queck, S.Y.; Koh, K.S.; Rice, S.A.; Givskov, M.; Kjelleberg, S. Quorum sensing-controlled biofilm development in Serratia liquefaciens MG1. J. Bacteriol. 2004, 186, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Domka, J.; Lee, J.; Wood, T.K. YliH (BssR) and YceP (BssS) regulate escherichia coli K-12 biofilm formation by influencing cell signaling. Appl. Environ. Microbiol. 2006, 72, 2449–2459. [Google Scholar] [CrossRef] [PubMed]
- Kirillina, O.; Fetherston, J.D.; Bobrov, A.G.; Abney, J.; Perry, R.D. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol. Microbiol. 2004, 54, 75–88. [Google Scholar] [CrossRef]
- Bobrov, A.G.; Kirillina, O.; Perry, R.D. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol. Lett. 2005, 247, 123–130. [Google Scholar] [CrossRef]
- Kim, Y.; Wang, X.; Ma, Q.; Zhang, X.S.; Wood, T.K. Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J. Bacteriol. 2009, 191, 1258–1267. [Google Scholar] [CrossRef]
- Ogasawara, H.; Yamamoto, K.; Ishihama, A. Role of the biofilm master regulator Csgd in cross-regulation between biofilm formation and flagellar synthesis. J. Bacteriol. 2011, 193, 2587–2597. [Google Scholar] [CrossRef]
- Labrie, J.; Pelletier-Jacques, G.; Deslandes, V.; Ramjeet, M.; Auger, E.; Nash, J.H.; Jacques, M. Effects of growth conditions on biofilm formation by Actinobacillus Pleuropneumoniae. Vet. Res. 2010, 41, 3. [Google Scholar] [CrossRef]
- Römling, U.; Galperin, M.Y.; Gomelsky, M. Cyclic Di-GMP: The first 25 Years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 2013, 77, 1–52. [Google Scholar] [CrossRef]
- Gerstel, U.; Park, C.; Römling, U. Complex regulation of Csgd promoter activity by global regulatory proteins. Mol. Microbiol. 2003, 49, 639–654. [Google Scholar] [CrossRef]
- Johnson, R.; Mylona, E.; Frankel, G. Typhoidal salmonella: Distinctive virulence factors and pathogenesis. Cell. Microbiol. 2018, 20, e12939. [Google Scholar] [CrossRef] [PubMed]
- Keitel, W.A.; Bond, N.L.; Zahradnik, J.M.; Cramton, T.A.; Robbins, J.B. Clinical and serological responses following primary and booster immunization with salmonella typhi vi capsular polysaccharide vaccines. Vaccine 1994, 12, 195–199. [Google Scholar] [CrossRef]
- Hu, X.; Chen, Z.; Xiong, K.; Wang, J.; Rao, X.; Cong, Y. Vi Capsular polysaccharide: Synthesis, virulence, and application. Crit. Rev. Microbiol. 2017, 43, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.M.; Baron, L.S. Genetic transfer of the Vi antigen from Salmonella typhosa to Escherichia coli. J. Bacteriol. 1969, 99, 358–359. [Google Scholar] [CrossRef] [PubMed]
- Wetter, M.; Goulding, D.; Pickard, D.; Kowarik, M.; Waechter, C.J.; Dougan, G.; Wacker, M. Molecular characterization of the ViaB locus encoding the biosynthetic machinery for Vi capsule formation in Salmonella Typhi. PLoS ONE 2012, 7, e45609. [Google Scholar] [CrossRef]
- Al Safadi, R.; Abu-Ali, G.S.; Sloup, R.E.; Rudrik, J.T.; Waters, C.M.; Eaton, K.A.; Manning, S.D. Correlation between in vivo biofilm formation and virulence gene expression in Escherichia coli O104:H4. PLoS ONE 2012, 7, e41628. [Google Scholar] [CrossRef]
- Bassler, B.L. How bacteria talk to each other: Regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 1999, 2, 582–587. [Google Scholar] [CrossRef]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef]
- Nealson, K.H.; Platt, T.; Hastings, J.W. Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 1970, 104, 313–322. [Google Scholar] [CrossRef]
- Seshadri, R.; Joseph, S.W.; Chopra, A.K.; Sha, J.; Shaw, J.; Graf, J.; Haft, D.; Wu, M.; Ren, Q.; Rosovitz, M.J.; et al. Genome sequence of Aeromonas hydrophila ATCC 7966T: Jack of all trades. J. Bacteriol. 2006, 188, 8272–8282. [Google Scholar] [CrossRef]
- Igbinosa, E.O.; Rathje, J.; Habermann, D.; Brinks, E.; Cho, G.S.; Franz, C.M.A.P. Draft genome sequence of multidrug-resistant strain Citrobacter portucalensis MBTC-1222, isolated from uziza (Piper guineense) leaves in Nigeria. Genome Announc. 2018, 6, e00123-18. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.S.; Sultana, M.; Hossain, M.A. Complete genome arrangement revealed the emergence of a poultry origin superbug Citrobacter portucalensis strain NR-12. J. Glob. Antimicrob. Resist. 2019, 18, 126–129. [Google Scholar] [CrossRef] [PubMed]
- Holmes, P.; Niccolls, L.M.; Sartory, D.P. The ecology of mesophilic Aeromonas in the aquatic environment. In The Genus: Aeromonas, 1st ed.; Austin, B., Altwegg, M., Gosling, P.J., Joseph, S.W., Eds.; John Wiley & Sons: Chicester, UK, 1996; p. 127. [Google Scholar]
- Martin-Carnahan, A.; Joseph, S.W. Aeromonas. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Brenner, D.J., Krieg, N.R., Staley, J.T., Garrity, G.M., Eds.; Williams and Wilkins: New York, NY, USA, 2005; Volume 2. [Google Scholar]
- Pasquale, V.; Baloda, S.B.; Dumontet, S.; Krovacek, K. An outbreak of Aeromonas hydrophila infection in turtles (Pseudemis scripta). Appl. Environ. Microbiol. 1994, 60, 1678–1680. [Google Scholar] [CrossRef] [PubMed]
- Xianle, Y.; Fuen, K.; Jianguang, Z.; Xiaohui, A. Virulence of Aeromonas hydrophila isolated from diseased soft-shelled turtle, Trionyx sinensis. J. Fish. Sci. China 1999, 6, 107–121. [Google Scholar]
- Gao, G.; Shi, Q.; Zhang, Y.; Gao, G.; Chen, C. Separation and identification of pathogens causing soft-shelled turtle fulminant infectious disease and preparation of anti-aeromonas serum. Agric. Sci. Technol. 2012, 13, 2155–2158. [Google Scholar]
- Shan, Q.; Zheng, G.; Liu, S.; Bai, Y.; Li, L.; Yin, Y.; Ma, L.; Zhu, X. Pharmacokinetic/pharmacodynamic relationship of marbofloxacin against Aeromonas hydrophila in Chinese soft-shelled turtles (Trionyx sinensis). J. Vet. Pharmacol. Ther. 2015, 38, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Delli Paoli Carini, A.; Ariel, E.; Picard, J.; Elliott, L. Antibiotic resistant bacterial isolates from captive green turtles and in vitro sensitivity to bacteriophages. Int. J. Microbiol. 2017, 2017, 5798161. [Google Scholar] [CrossRef]
- Palma-Martínez, I.; Guerrero-Mandujano, A.; Ruiz-Ruiz, M.J.; Hernández-Cortez, C.; Molina-López, J.; Bocanegra-García, V.; Castro-Escarpulli, G. Active Shiga-like toxin produced by some Aeromonas spp., isolated in Mexico City. Front. Microbiol. 2016, 7, 1522. [Google Scholar] [CrossRef]
- Grotiuz, G.; Sirok, A.; Gadea, P.; Varela, G.; Schelotto, F. Shiga toxin 2-producing Acinetobacter haemolyticus associated with a case of bloody diarrhea. J. Clin. Microbiol. 2006, 44, 3838–3841. [Google Scholar] [CrossRef]
- Torres, A.G.; Jeter, C.; Langley, W.; Matthysse, A.G. Differential binding of Escherichia coli O157:H7 to alfalfa, human epithelial cells, and plastic is mediated by a variety of surface structures. Appl. Environ. Microbiol. 2005, 71, 8008–8015. [Google Scholar] [CrossRef]
- Raad, I.; Costerton, W.; Sabharwal, U.; Sacilowski, M.; Anaissie, E.; Bodey, G.P. Ultrastructural analysis of indwelling vascular catheters: A Quantitative relationship between luminal colonization and duration of placement. J. Infect. Dis. 1993, 168, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Stickler, D.; Morris, N.; Moreno, M.C.; Sabbuba, N. Studies on the formation of crystalline bacterial biofilms on urethral catheters. Eur. J. Clin. Microbiol. Infect. Dis. 1998, 17, 649–652. [Google Scholar] [CrossRef] [PubMed]
- Norton, T.; Thompson, R.; Pope, J.; Veltkamp, C.; Banks, B.; Howard, C.; Hawkins, S. Using confocal laser scanning microscopy, scanning electron microscopy and phase contrast light microscopy to examine marine biofilms. Aquat. Microb. Ecol. 1998, 16, 199–204. [Google Scholar] [CrossRef]
- Hannig, C.; Follo, M.; Hellwig, E.; Al-Ahmad, A. Visualization of adherent micro-organisms using different techniques. J. Med. Microbiol. 2010, 59, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Gagné-Thivierge, C.; Barbeau, J.; Levesque, R.C.; Charette, S.J. A new approach to study attached biofilms and floating communities from Pseudomonas aeruginosa strains of various origins reveals diverse effects of divalent ions. FEMS Microbiol. Lett. 2018, 365, fny155. [Google Scholar] [CrossRef] [PubMed]
- Armbruster, C.R.; Parsek, M.R. New insight into the early stages of biofilm formation. Proc. Natl. Acad. Sci. USA 2018, 115, 4317–4319. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).