Abstract
Weissella cibaria is one of the bacteria in charge of the initial fermentation of kimchi and has beneficial effects such as immune-modulating, antagonistic, and antioxidant activities. In our study, we aimed to estimate the safety of W. cibaria JW15 for the use of probiotics according to international standards based on phenotypic (antibiotic resistance, hemolysis, and toxic metabolite production) and genotypic analysis (virulence genes including antibiotic resistance genes). The results of the safety assessment on W. cibaria JW15 were as follows; (1) antibiotic resistance genes (ARGs) (kanamycin and vancomycin etc.) were intrinsic characteristics; (2) There were no acquired virulence genes including Cytolysin (cylA), aggregation substance (asa1), Hyaluronidase (hyl), and Gelatinase (gelE); (3) this strain also lacked β-hemolysis and the production of toxic metabolites (D-lactate and bile salt deconjugation). Consequently, W. cibaria JW15 is expected to be applied as a functional food ingredient in the food market.
1. Introduction
In 1965, “probiotics” were first described as growth-promoting factors produced by microorganisms [1] and current probiotics are defined as ‘live micro-organisms which when administered in adequate amounts confer a health benefit on the host’ by the Food and Agriculture Organization of the United Nations and the World Health Organization (FAO/WHO) [2]. This characteristic of probiotics has been observed in bacteria, yeast, and fungi, but commonly used probiotics belong to lactic acid bacteria (LAB) and bifidobacteria, and their species are as following Lactobacillus (L. acidophilus, L. gasseri, L. delbrueckii subsp. bulgaricus, and L. helveticus), Lacticaseibacillus (L. casei, L. paracasei, and L. rhamnosus), Limosilactobacillus (L. fermentum and L. reuteri), Lactiplantibacillus (L. plantarum), Ligilactobacillus (L. salivarius), Lactococcus (Lc. lactis), Enterococcus (E. faecium and E. faecalis), Streptococcus (S. thermophilus), and Bifidobacterium (B. bifidum, B. breve, B. longum, and B. animalis subsp. lactis) [3,4].
Commercial starter culture products have been constantly consumed in the fermented food market as microbial food cultures (MFC) including LAB. Probiotic strains of LAB are also used in diverse medical and health-related areas, including the treatment of infections during pregnancy; management of allergic diseases; alleviation of intestinal inflammation; halt of antibiotic-related diarrhea, and prevention of urinary tract infections [5]. Of these, several probiotics such as L. rhamnosus GG (LGG), B. animalis subsp. lactis BB-12, and L. casei Shirota, etc. have been developed by global companies in the field of probiotics and they have also recently been marketed in the form of tablets or powders [6,7].
Probiotics are well-known and generally classified as safe (GRAS) because of their long-term safety in dairy products or fermented foods. The Lactic Acid Bacteria Industrial Platform (LABIP) has reported that the risk of infection caused by LAB occurs very rarely except for enterococci and streptococci [7]. However, in recent years, many controversies have been raised over the safety of probiotics since bacteria used in probiotics are frequently isolated from infection sources [3]. It should be noted that not all LAB of a particular genus or species have probiotic properties and are assigned to a particular strain such as Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus salivarius subsp. thermophilus [8].
To evaluate the safety of probiotics, guidelines that take into account several factors in advance, such as excessive immune stimulation in sensitive individuals, systemic infection, gene transfer, or deleterious metabolic effects, are needed [9]. In 2002, FAO/WHO notes that it is important to conduct safety assessments, including production of certain metabolites such as D-lactate and ammonia, adverse effects in humans, antibiotic resistance, potential hemolysis, and toxin production, even for microorganisms classified as GRAS [2].
Weissella sp. is an LAB with the phenotypic properties of being Gram-positive, non-spore-forming, non-motile, etc. The genus Weissella is detected in various natural environments such as fermented foods (kimchi, fermented fava-bean, sausages etc.) and digestive tracts of humans and animals [10,11]. Many studies have shown that W. cibaria, which appears pro-dominant in the initial fermentation of kimchi, has beneficial effects such as probiotic properties, antimicrobial-, antagonistic-, and antioxidant-activities, etc. [12,13,14]. In addition to animal trials, the immunomodulatory activity of W. cibaria JW15 was significantly higher than that of L. rhamnosus GG, a well-known immune enhancer [12]. Recently, W. cibaria has been registered as a safe raw material by the Korea Food and Drug Administration (KFDA) [4] and is actively commercialized as a food ingredient in Korea. In 2018, the International Dairy Federation (IDF) is suggesting the use of W. cibaria as the Microbial Food Cultures (MFC) for the food usage of vegetables [15].
This study aimed to verify the safety of W. cibaria JW15 according to the international standards of FAO/WHO based on phenotypic (antibiotic resistance, hemolysis, and toxic metabolite production) and genotypic analysis (virulence genes including antibiotic resistance genes). Furthermore, W. cibaria JW15 was evaluated by the bacterial reverse mutation to identify genotoxicity.
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
In our study, we used three Weissella cibaria strains for safety evaluation. Based on a previous study, Weissella cibaria JW15 (registered as KACC 91811P, Accession: NZ_CP-CP058237-CP058240), which is isolated from Kimchi, was selected as a bacterium that had immune-enhancing properties through NK cell activation [14]. Lactobacillus rhamnosus GG (ATCC 53103), W. cibaria LMG 21843, and W. cibaria LMG 17699 were purchased from the American Type Culture Collection (Manassas, VA, USA) and the BCCM/LMG Bacteria Collection (Ghent, Belgium) and used as reference strains. The W. cibaria strains and L. rhamnosus GG were cultivated anaerobically in De Man, Rogosa, and Sharpe broth (MRS broth, Merk, Darmstadt, Germany) at 30 °C for 24 h.
2.2. Minimum Inhibitory Concentrations on Antibiotics
The minimum inhibitory concentration (MIC) was determined using a commercial E-test (Epsilometer test, bioMerieux, France): Ampicillin, Gentamicin, Kanamycin, Streptomycin, Erythromycin, Clindamycin, Tetracycline, and Chloramphenicol. The concentration on the strips ranged from 0.016 to 256 μg/mL except for streptomycin (0.064 to 1024 μg/mL). Bacterial suspensions were adjusted to a turbidity of 0.5 of the McFarland standard (bioMerieux, Marcy l’Etoile, France). The suspensions were inoculated using a sterile cotton swab on the entire surface of the MRS agar plate. E-test strips were placed on the surface of the inoculated agar and anaerobically incubated at 35 °C for 48 h. The MIC was interpreted as the point at which the ellipse intersected the E-test strip according to the manufacturer’s instructions.
2.3. Detection of Antibiotic Resistance Genes
Chromosomal DNA was extracted from the bacteria using an AllPrep® Bacterial DNA/RNA/Protein Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instruction manuals. Plasmid DNA was also extracted from 2 mL of bacterial culture using an EzPureTM Plasmid prep kit (Enzynomics, Inc, Daejeon, ROK) according to the manufacturer’s instructions with the cell lysis buffer (20 mM Tris-Cl, pH 8.0, 2 mM sodium EDTA, and 1.2% Triton X-100 containing 20 mg/mL lysozyme). The quantity of DNA was assessed with a Nanodrop spectrophotometer (2000, Thermo Fisher Scientific, Waltham, MA, USA). The chromosomal and plasmid DNA extracts from the strains were diluted and used for polymerase chain reactions (PCRs) to detect ARGs targeted by gene-specific primers [16,17,18,19,20]. The PCR was performed with an initial denaturation step of 95 °C for 5 min, followed by 35 cycles of 95 °C for 10 s, annealing temperature (Table 1) for 30 s, 72 °C for 60 s. To confirm the amplicons of ARGs, PCR products were loaded into in agarose gels for electrophoresis. The primer sequences are listed in Table 1.

Table 1.
Primer and gene sequences used in this study.
2.4. Genomic Analysis
We performed the genomic analysis of the JW15 strain using Macrogen service (Macrogen Inc., Seoul, Korea). The manufacturer’s instructions were as follows; DNA samples were sequenced using the PacBio RS II platform and Illumina HiseqXten platform, and then the subheads generated from PacBio RS II were assembled using the hierarchical genome assembly process (HGAP) [21] with default options. For error correction, the Illumina raw reads were filtered by quality at a level of 90% of bases had a phred score of 30 or higher. The assembly was corrected using high-quality HiseqXten reads by Pilon v1.21 [22]. Prokka v1.13 [23] was used for gene prediction and basic annotation. For additional annotation, the predicted protein sets were subjected to InterProScan v5.30-69.0 [24] and psiblast v2.4.0 [25] with EggNOG DB v4.5 [26]. Circular maps depicting each contig were generated using Circos v0.69.3 [27].
2.5. Bioinformatic Analysis of Virulence Factor-Related Genes
The VF-and toxin genes in the genome of the strain JW15, including Contig 1 to 4, were searched through BlastX analysis using Diamond software (ver. 0.9.26.127) [28] based on the virulence factor database (VFDB, http://www.mgc.ac.cn/VFs/, accessed on 12 August 2021) [29], which is an integrated and comprehensive online resource for curating information about virulence factors of bacterial pathogens. Thresholds for percent identity (% ID) and minimum length were set at 50% and 70%, respectively. In detail, VF-related genes, including those associated with enterotoxin, leukotoxin, cytolysin, cytotoxin K, hemolysis, biogenic amine production, hyaluronidase, aggregation, enterococcal surface protein, endocarditis antigen, collagen adhesion, cereulide, sex pheromone, and serine protease. These genes were additionally confirmed through BlastX analysis using experimentally-verified VF and toxin genes in the UniRef90 database.
2.6. Hemolytic Activity
W. cibaria strains, LGG, and Bacillus cereus KACC 10004 were used as positive controls for hemolytic activity. The strains were aerobically cultured in blood agar supplemented with 5% sheep blood at 37 °C for 2 days. The plates were then analyzed for microbial hemolytic properties by illuminating and observing the plate. Colonies that revealed green-hue zones (α-hemolysis) or did not reveal any hemolysis (γ-hemolysis) were considered non-hemolytic strains. Colonies that displayed blood lyses zones (clear zones) were classified as hemolytic (β-hemolysis) strains.
2.7. D-Lactic Acid Measurement
The production of D-lactic acid by W. cibaria strains and LGG was measured using the d-lactate colorimetric assay kit from BioVision Research (Mountain View, CA, USA). The LAB strains were cultured in MRS broth for 24 h at 37 °C, and the supernatant was used for this experiment.
2.8. Bile Salt Deconjugation
Bile salt deconjugation was carried out according to the plate assays of Dashkevicz and Feifhner [30]. W. cibaria strains and LGG were cultured for 24 h at 37 °C on an MRS agar plate containing 0.5% taurodeoxycholic acid (TDCA; Sigma, St. Louis, MO, USA). The results were interpreted as positive in the case of the formation of a halo of sediment or opaque granular white colonies around the colonies.
2.9. Enzymatic Profiles by API ZYM
Use of the API ZYM kit (bioMérieux, Marcy l’Etoile, France) was based on a substrate availability of a total of 19 enzymes. The bacterial suspension was adjusted with McFarland no. 5 being dropped in each tube. After incubation at 37 °C for 4 h, the results were determined to be positive if the color intensity was more than three following the manufacturer’s instructions.
2.10. Bacterial Reverse Mutation Assay
A bacterial reverse mutation assay was performed to evaluate the mutagenicity of W. cibaria JW15 with or without the S9 mix, following the principles of OECD Guideline 471 (2020) [31]. The assay was carried out using Salmonella typhimurium histidine-auxotrophic strains TA98, TA100, TA1535, TA1537, and Escherichia coli tryptophan-auxotrophic strain WP2uvrA (Molecular Toxicology, Boone, INC, USA). The S9 mix was used as a metabolic activation system (ORIENTAL YEAST Co., Ltd., Tokyo, Japan) and was prepared at the time of use in the required amount. Freeze-stored S9 (Lot No.: 20121110) and Cofactor A (Lot No.: A20120810) were thawed and prepared by mixing at a ratio of 1:9. Different dilutions of W. cibaria JW15 samples (5000, 2500, 1250, 625, and 313 μg/plate) were used for all tests under the same conditions. After being cultured at 37 °C for 48 h, the number of colonies in each tested group was counted per plate. This result was determined to be positive when the revertant colonies in the subject group were more than doubled. The data of historical control is presented in Table S1 as Supplementary Information.
3. Results and Discussion
The aim of this study was to verify the safety of W. cibaria JW15 based on phenotypic (antibiotic resistance, hemolysis, and toxic metabolite production) and genotypic analyses (virulence genes including antibiotic resistance genes). Currently, W. cibaria has no use as a probiotic ingredient, and the species is reported on antibiotic resistance such as kanamycin and vancomycin. Nevertheless, they have been frequently isolated from fermented foods and human feces and are well-known for their beneficial effects such as probiotic properties, antimicrobial-, antagonistic-, and antioxidant activities etc. Many researchers or consumers expect higher functional or healthy foods made from lactic acid bacteria with novel activity.
3.1. Determination of Minimum Inhibitory Concentrations
Weissella spp. has not been cleared on the cut-off values of MIC against antibiotics by EFSA in 2012. Accordingly, we determined an antibiotic susceptibility test of the W. cibaria JW15 strain corresponding to a Leuconostoc spp., based on the EFSA cut-off value, which reflects the phylogenetic and phenotypic characterization of the JW15 strain.
To ensure safety, the phenotypic antibiotic susceptibility of W. cibaria JW15 was investigated against 9 antibiotics, including ampicillin (AM), chloramphenicol (CL), clindamycin (CM), erythromycin (EM), gentamicin (GM), kanamycin (KM), streptomycin (SM), tetracycline (TC), and vancomycin (VA) using the E-test method [32]. As shown in Table 2, W. cibaria JW15 was susceptible to 7 kinds of antibiotics, including ampicillin (AM), chloramphenicol (CL), clindamycin (CM), erythromycin (EM), gentamicin (GM), streptomycin (SM), and tetracycline (TC), which were found below the cut-off value (μg/mL) within the safe range. However, the JW 15 strain was shown to be resistant to kanamycin (KM).

Table 2.
Antibiotic resistance profiles and minimum inhibitory concentration (MIC) values of bacterial strains used in this study.
Recent studies have shown that the antibiotic susceptibility profile of W. cibaria differs between each strain [33,34]. W. cibaria CMU was found to be sensitive to AM, CL, CM, EM, GM, SM, and TC, except for KM corresponding to an obligate hetero-fermentative lactobacilli [33]. In our result, W. cibaria strains showed MICs ≥ 256 mg/L for kanamycin and vancomycin, suggesting that the resistance against kanamycin and vancomycin could be considered an intrinsic property. Antibiotic resistance was found not only in the genus Weissella, but also in many lactic acid bacteria used as food ingredients. Lactobacillus sp. shows high resistance to antibiotics reported as endogenous with strong resistance to antibiotics such as kanamycin and vancomycin [17,34]. It has been reported that lactic acid bacteria derived from fermented food are resistant to antibiotics [35,36]. Therefore, it has been speculated that the characteristic that the JW15 strain isolated from kimchi is resistant to some antibiotics may be common.
3.2. Detection of Antibiotic Resistance Genes
The transferability of antibiotic resistance (AR) genes and plasmids present in bacteria is associated with human health. Here, we confirmed the existence of AR genes and plasmids in the W. cibaria JW15 strain on four antibiotics (clindamycin, kanamycin, streptomycin, and vancomycin) showing high MIC cut-off value presented in Table 2.
PCR analysis for four antibiotic resistance genes such as streptomycin (aadA, aadE, and strB), tetracycline (tet (K)), kanamycin (aph (3”)-III and ant (2”)-I), and clindamycin (Inu (A) and Inu (B)) were conducted. Although there was detected amplicons in several samples, they were not antibiotic resistant genes based on sequencing analysis. The PCR results are shown in Table 3. There was no expected amplicon in the chromosome and plasmid DNA of W. cibaria JW15, W. cibaria LMG 21843, W. cibaria LMG 17699, and L. rhamnosus ATCC 53103 used in this study.

Table 3.
Detection for antibiotics resistance genes (ARGs).
For antibiotic resistance, the MIC cut-off value of kanamycin was exceeded, which is a phenotypic evaluation, but the antibiotic resistance target gene was not detected in the chromosome and plasmid of the JW15 strain, which is a genotype evaluation. Sharma et al. (2014) reported that antibiotics intrinsic strains were phenotypically resistant may be genotypically susceptible [37]. We found several studies showing this characteristic, and strains that also had specific antibiotic resistance, but no gene was detected [33,38]. Therefore, the results of antibiotic resistance to KM and detection of their target genes are similar to those seen in antibiotic intrinsic strains according to previous reports. In addition, the phenotypic property of the JW15 strain that exhibits resistance to kanamycin may be due to four endogenous-related mechanisms such as enzyme inactivation or modification, alteration of bacterial target sites, antibiotic efflux pump and outer membrane permeability change, and intracellular metabolic rearrangement [37].
Moreover, for the transferability of antibiotic resistance, the plasmid plays a major role in the ARG gene transfer method (HGT) [37]. In our results, as shown in Table 3, kanamycin resistance gene (aph (3″)-III and ant (2″)-I) were not detected in the plasmid of JW15, thus the transferability is considered low.
3.3. Genomic Features of JW15 Strain
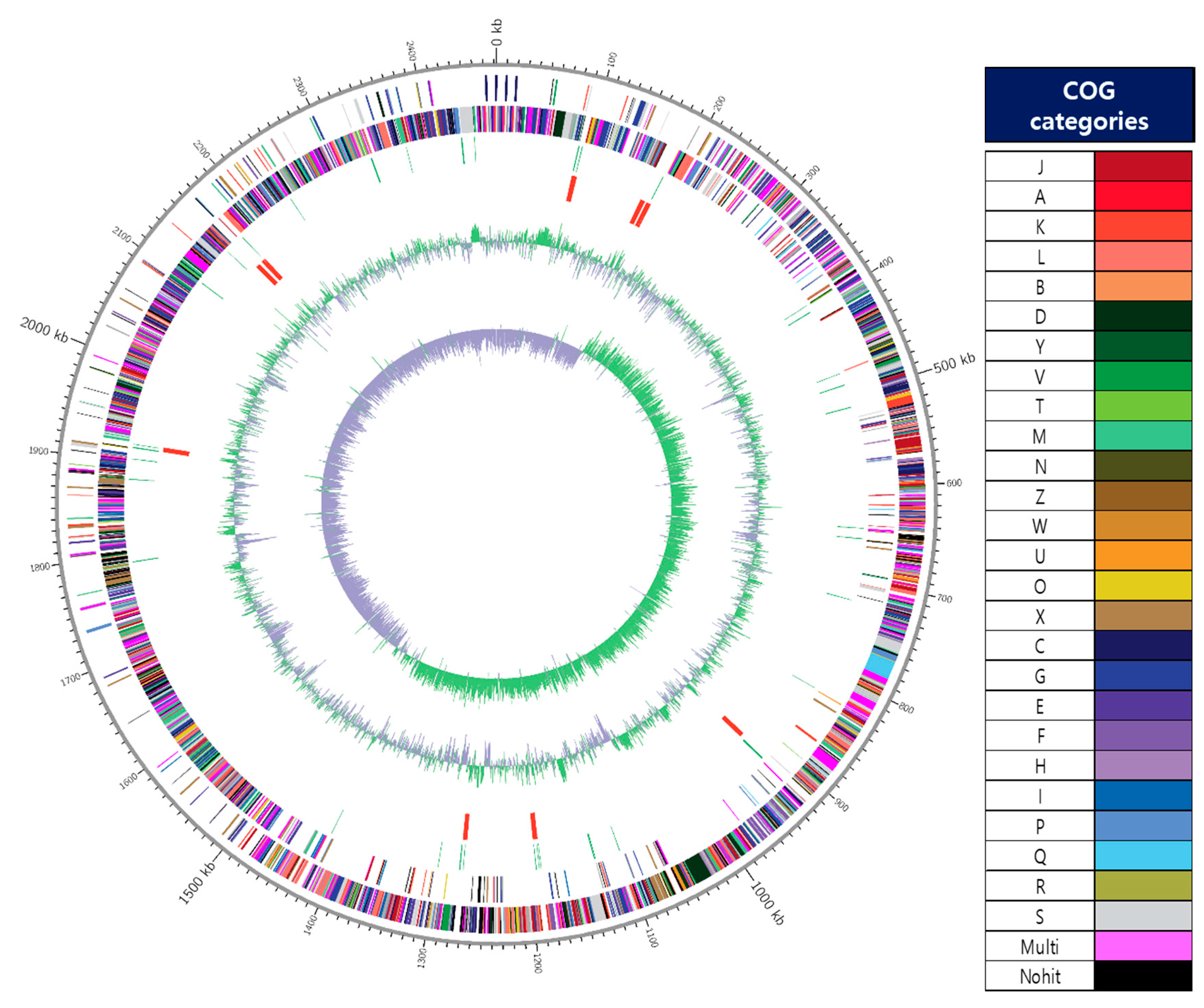
The key genomic features of W. cibaria JW15, including GC skew, protein-coding sequences (CDSs), COG categories, and G+C contents, are graphically depicted in Figure 1. The genome of the JW15 strain was a single circular chromosome of 2,472,214 bp with 3 plasmids (30,944 bp, 17,267 bp, and 14,411 bp). The genome of strain JW15 contains a total of 2315 CDS, 42 tRNA genes, and 28 rRNA genes. The result of COG-assigned proteins in the genomes of strain JW15 and their distributions into COG categories was not abbreviated. As a result, the COGs were classified into 26 functional categories except for Nohit against the COG database and of the 2556 protein-coding genes, 2259 genes (88.39%) were assigned to COGs categories. The W. cibaria UTNGt21O strain (1635 genes) reported by Tenea and Hurtado [39] was less than the W. cibaria JW15 strain. We found that the essential genes from the functional subcategories with the COG codes G (Carbohydrate transport and metabolism, 7.75%), J (Translation, ribosomal structure, and biogenesis, 7.67%), K (Transcription, 5.95%), L (Replication, recombination and repair, 4.38%), H (Coenzyme transport and metabolism, 3.44%), and I (Lipid transport and metabolism, 3.4%). However, the distribution of functional annotation of W. cibaria UTNGt21O strain was differently expressed in the order of R (General function prediction only, 8.99%), J (Translation, ribosomal structure, and biogenesis, 8.07%), K (Transcription, 6.60%), and L (Replication, recombination and repair, 6.54%) in comparison with W. cibaria JW15 strain. Owing to different genes, depending on strain-specificity, the information of functional genes mentioned here will help additional studies of this strain and demonstrate its potential property for the use of probiotics.
Figure 1.
A circular map of the chromosome of JW15 strain. The map was drawn by applying Contig 1’s annotation result. From outside to the center: coding sequences (CDS) on forward strand (colored by COG categories of the right side), CDS on reverse strand (colored by COG categories of the right side.), tRNA, rRNA, GC content, and GC skew (+: green, −: violet). The complete genome contained 2,472,214 bp with G+C content of 45.09%.
3.4. Bioinformatic Analysis of VF-Related Genes
In our study, we did not discover virulence-related genes in the chromosomal and plasmid genomes of the W. cibaria JW15 strain as shown in Table 4. However, two genes in JW15_contig1 (Table 5) were identified with low homology (loose identity, <95%) with the virulence factor database (VFDB) containing information on the virulence genes of bacterial pathogens online. Gene JW15-00598 showed a homology of 57.4% to gene efaA involved in endocarditis antigen, and gene JW15-00853 showed a homology of 53.1% to gene CD1208 (or CVF417) involved in RNA methyltransferase (or hemolysin A). In addition, the two genes (gene JW15-00598 and JW15-00853) researched in NCBI and Uniprot were found to have high homology (>95%) with transporter substrate-binding protein, RNA methyltransferase or cell division, respectively. In particular gene JW15-00853, which was identified as TlyA, is known to be not, on its own, a potent hemolysin [40]. Therefore, the two genes are presumed to be general transporters and transferase genes that are not related to toxic genes such as endocarditis antigen and hemolysin. In addition, the W. cibaria JW15 strain was negative in the hemolysis test, which was consistent with the bioinformatic analysis. The gene sequences are shown in Table S1 in Supplementary Materials. The VF-related gene information that was used to confirm the safety of the strain should help further probiotic studies.

Table 4.
Bioinformatic analysis for the presence of putative virulence factor-related genes in the genomes of strain JW15.

Table 5.
Homology analysis of two genes presumed to be VF-genes in the genome of W. cibaria JW 15 strain.
3.5. Toxic Metabolite Production
3.5.1. Hemolytic Activity
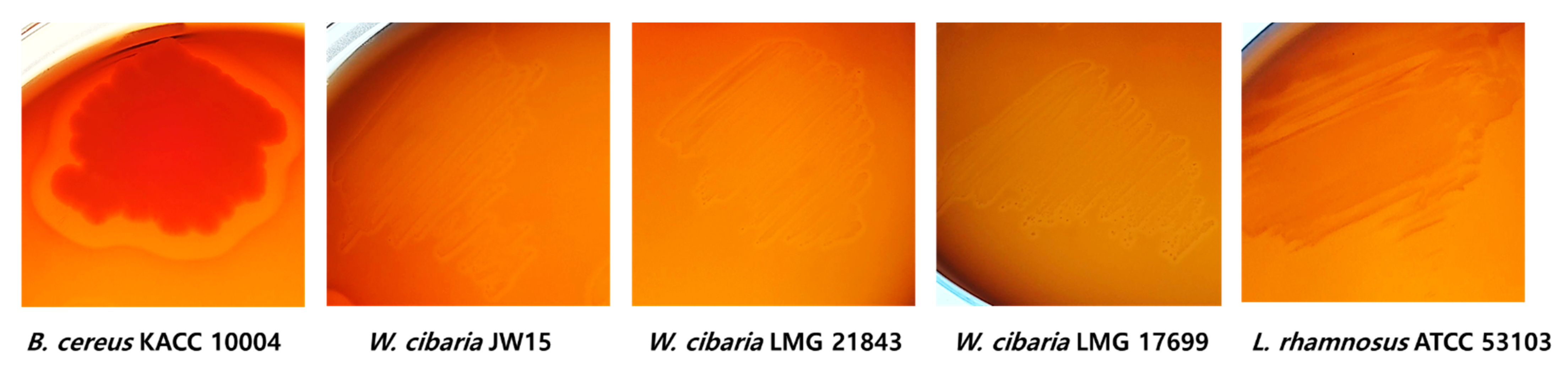
Hemolysin is a toxic enzyme of pathogenic bacteria such as Bacillus cereus and has hemolytic activity to destroy red blood cells in the host, as well as the possibility for edema and anemia [38,41]. Generally, β-hemolysis is associated with microbial pathogenicity. In our study, B. cereus KACC 10004 as a positive control showed clear zones (expressed as β-hemolysis) around the colonies, whereas W. cibaria strains and LGG did not show β-hemolysis activity (Figure 2).
Figure 2.
Hemolytic activity of W. cibaria strains and LGG. Complete lysis of blood cells was observed, with clear zones around B. cereus KACC 10004 as positive control.
3.5.2. D-Lactic Acid Production
Various bacterial species are known to produce D-lactate or both D- and L-lactates are produced in fermentation. Of them, the genus Lactobacillus produces D- and L-lactates, the genus Pediococcus produces L-, and the genera Leuconostoc, Oenococcus, and Weissella produce D-lactic acid [42]. As shown in Table 6, the productivity of D-lactic acid by W. cibaria was measured by enzymatic assays concerning D-lactate dehydrogenase. W. cibaria strains did not produce D-lactic acid like the commercial probiotic strain LGG. Our result was similar to the report showing that the W. cibaria CMU strain was unable to produce D-lactic acid [33].

Table 6.
Enzymatic profiles and assay of toxic metabolic production.
3.5.3. Bile Salt Deconjugation Test
Bile salts are less capable of solubilizing and absorbing lipids in the gut. All strains used in this study were able to grow in the presence or absence of sodium taurodeoxycholate (0.5%) and did not show the precipitate halos or the opaque white colonies after growth in MRS with TDCA (Table 6). These results show the lack of ability to deconjugate sodium taurodeoxycholate and agree that W. cibaria could not convert to secondary bile acid as previously published report [36].
3.5.4. Enzymatic Profile by API ZYM
The enzyme profile of the JW15 strain was similar to that of the LMG 28143 strain isolated from fermented kimchi, while the LMG 17699 strain was different from the β-galactosidase β-glucosidase enzymes (Table 6). In Muñoz-Atienza et al. (2013), it was observed that leucine arylamidase, valine arylamidase, β-galactosidase, and β-glucosidase showed different patterns among the 15 kinds of Weissella spp. [36]. In the case of the β-glucuronidase, no generation of potential carcinogenic metabolites [43] was detected in any of the W. cibaria strains and LGG, indicating that they are safe.
3.6. Bacterial Reverse Mutation Assay
The bacterial reverse mutation assay, developed by Bruce Ames in 1973 [44], was performed for mutagenicity testing of probiotics such as L. rhamnosus, B. adolescentis, L. paracasei, L. mali, and P. acidilactici [45,46,47]. The genotoxicity was conducted by this assay with different doses of W. cibaria JW15 against four mutant S. typhimurium strains (TA98, TA100, TA1535, and TA1537) and a mutant E. coli strain (WP2uvrA), respectively. An expected increase of revertant colonies was observed in all positive groups after induction of the mutants. After exposure of bacterial strain to different concentrations of W. cibaria JW15, the number of revertant colonies, regardless of the presence or absence of S9 mix, did not exceed twice that of the negative control group (Table 7). Therefore, the W. cibaria JW15 treatment groups were considered not to have mutagenic activity in the histidine auxotrophy of the S. typhimurium strains or the tryptophan auxotrophy of E. coli.

Table 7.
Mutagenic activity in bacterial strains TA98, TA100, TA1535, TA1537, and WP2uvrA treated with W. cibaria JW15, with (+S9) or without (−S9) metabolic activation.
Consequently, in this study, we verified the safety of the W. cibaria JW15 strain by phenotypic and genotypic property analysis according to the international guidelines by FAO/WHO. The safety was evaluated by a minimum inhibitory concentration assay for 9 antibiotics, chromosomal and plasmid DNA analysis for 12 antibiotic resistance genes (ARGs) on 4 antibiotics, virulence gene analysis, beta-hemolysis, toxic metabolite production, and bacterial reverse mutation assay. The strain W. cibaria JW15 was susceptible to all antibiotics except for kanamycin and vancomycin. We confirmed that there was no harboring of antibiotic resistance target genes and virulence-related genes in the genome of strain JW15. We therefore considered that antibiotic resistance (e.g., kanamycin, vancomycin) was an intrinsic property of W. cibaria JW15. Additionally, the strain JW15 lacked β-hemolysis, β-glucuronidase, toxic metabolites such as D-lactate and bile salt deconjugation, and bacterial reverse mutagenic activity. Accordingly, we believe that W. cibaria JW15 could be commercially applied as a probiotic strain in the future.
Supplementary Materials
The following are available online at www.mdpi.com/article/10.3390/microorganisms9122450/s1, Table S1: Data of historical positive/negative controls.
Author Contributions
Conceptualization, S.-Y.K.; methodology, Y.-J.J.; validation, S.-Y.K.; formal analysis, W.-S.J.; investigation, H.-M.G.; writing—original draft preparation, Y.-J.J.; writing—review and editing, S.-Y.K.; visualization, Y.-J.J.; supervision, S.-Y.K.; project administration, S.-H.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Research Program for Agricultural Science & Technology Development (Project No. PJ01416104) and the National Institute of Agricultural Sciences, Rural Development Administration, Republic of Korea.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
This genome sequence was deposited in GenBank (BioProject number PRJNA639573; and GenBank accession numbers CP058237-CP058240). The version described in this paper is the first version.
Acknowledgments
The financial support of the Rural Development Administration is gratefully acknowledged.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lilly, D.M.; Stillwell, R.H. Probiotics: Growth-promoting factors produced by microorganisms. Science 1965, 147, 747–748. [Google Scholar] [CrossRef]
- Food and Agriculture Organization-World Health Organization (FAO/WHO). Report on Joint FAO/WHO Guidelines for the Evaluation of Probiotics in Food. 2002. Available online: https://www.who.int/foodsafety/fs_management/en/probiotic_guidelines.pdf (accessed on 4 April 2019).
- Anna, Z.-R.; Tyski, S. Are Probiotic Really Safe for Humans? Pol. J. Microbiol. 2018, 67, 251–258. [Google Scholar]
- List of Raw Materials Available for Food. Available online: https://www.foodsafetykorea.go.kr/foodcode/01_03.jsp?idx=12135 (accessed on 4 April 2019).
- Bernardeau, M.; Vernoux, J.P.; Henri-Dubernet, S.; Gueguen, M. Safety assessment of dairy microorganisms: The Lactobacillus genus. Int. J. Food Microbiol. 2008, 126, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Global Market Insights Inc. Probiotics Market Size to Exceed USD 64 Billion by 2023: Global Market Insights Inc. Available online: https://www.prnewswire.com/news-releases/the-global-probiotics-market-size-isexpected-to-reach-usd-66-03-billion-by-2024--300726946.html (accessed on 4 April 2019).
- Liong, M.T. Safety of probiotics: Translocation and infection. Nutr. Rev. 2008, 66, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishibashi, N.; Yamazaki, S. Probiotics and safety. Am. J. Clin. Nutr. 2001, 73, 465–470. [Google Scholar] [CrossRef]
- Lee, K.-W.; Park, J.-Y.; Chun, J.-Y.; Han, N.-S.; Kim, J.-H. Importance of Weissella Species during Kimchi Fermentation and Future Works. Microbiol. Biotechnol. Lett. 2010, 38, 341–348. [Google Scholar]
- Rizzello, C.G.; Coda, R.; Wang, Y.; Verni, M.; Kajala, I.; Katina, K.; Laitila, A. Characterization of indigenous Pediococcus pentosaceus, Leuconostoc kimchii, Weissella cibaria and Weissella confusa for faba bean bioprocessing. Int. J. Food. Microbiol. 2019, 302, 24–34. [Google Scholar] [CrossRef]
- Ahn, S.-B.; Park, H.-E.; Lee, S.-M.; Kim, S.-Y.; Shon, M.-Y.; Lee, W.-K. Characteristics and immuno-modulatory effects of Weissella cibaria JW15 isolated from Kimchi, Korea traditional fermented food, for probiotic use. J. Biomed. Res. 2013, 14, 206–211. [Google Scholar] [CrossRef]
- Yu, H.S.; Lee, N.K.; Choi, A.J.; Choe, J.-S.; Bae, C.H.; Paik, H.-D. Antagonistic and antioxidant effect of probiotic Weissella cibaria JW15. Food. Sci. Biotechnol. 2019, 28, 851–855. [Google Scholar] [CrossRef]
- Lee, Y.J.; Lee, A.; Yoo, H.J.; Kim, M.; Noh, G.M.; Lee, J.H. Supplementation with the probiotic strain Weissella cibaria JW15 enhances natural killer cell activity in nondiabetic subjects. J. Funct. Foods 2018, 48, 153–158. [Google Scholar] [CrossRef]
- Bourdichon, F.; Casaregola, S.; Farrokh, C.; Frisvad, J.C.; Gerds, M.L.; Hammes, W.P. Food fermentations: Microorganisms with technological beneficial use. Int. J. Food Microbiol. 2012, 154, 87–97. [Google Scholar] [CrossRef]
- Ouoba, L.I.; Lei, V.; Jensen, L.B. Resistance of potential probiotic lactic acid bacteria and bifidobacteria of african and european origin to antimicrobials: Determination and transferability of the resistance genes to other bacteria. Int. J. Food. Microbiol. 2008, 121, 217–224. [Google Scholar] [CrossRef]
- Kastner, S.; Perreten, V.; Bleuler, H.; Hugenschmidt, G.; Lacroix, C.; Meile, L. Antibiotic susceptibility patterns and resistance genes of starter cultures and probiotic bacteria used in food. Syst. Appl. Microbiol. 2006, 29, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, Z.Y.; Dong, K.; Yuan, J.P.; Guo, X.K. Antibiotic resistance of probiotic strains of lactic acid bacteria isolated from marketed foods and drugs. Biomed. Environ. Sci. 2009, 22, 401–412. [Google Scholar] [CrossRef]
- Aquilanti, L.; Garofalo, C.; Osimani, A.; Silvestri, G.; Vignaroli, C.; Clementi, F. Isolation and molecular characterization of antibiotic-resistant lactic acid bacteria from poultry and swine meat products. J. Food Prot. 2007, 70, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Aristimuño Ficoseco, C.; Mansilla, F.I.; Maldonado, N.C.; Miranda, H.; Fátima Nader-Macias, M.E.; Vignolo, G.M. Safety and Growth Optimization of Lactic Acid Bacteria Isolated from Feedlot Cattle for Probiotic Formula Design. Front. Microbiol. 2018, 9, 2220. [Google Scholar] [CrossRef]
- Chin, C.S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016, 44, D286–D293. [Google Scholar] [CrossRef] [Green Version]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using 447 DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dashkevicz, M.P.; Feifhner, S.D. Development of a Differential Medium for Bile Salt Hydrolase-Active Lactobacillus spp. Appl. Environ. Microbial. 1989, 55, 11–16. [Google Scholar] [CrossRef] [Green Version]
- OECD. OECD Guideline for the Testing of Chemicals 471, Bacterial Reverse Mutation Test; OECD: Paris, France, 1997. [Google Scholar]
- FAO; WHO. Probiotics in Food: Health and Nutritional Properties and Guidelines for Evaluation; World Health Organization; Food and Agriculture Organization of the United Nations: Rome, Italy, 2006. [Google Scholar]
- Kang, M.-S.; Yeu, J.-E.; Hong, S.-P. Safety Evaluation of Oral Care Probiotics Weissella cibaria CMU and CMS1 by Phenotypic and Genotypic Analysis. Int. J. Mol. Sci. 2019, 20, 2693. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wei, X.; Fan, M. Assessment of Antibiotic Susceptibility within Lactic Acid Bacteria and Coagulase-Negative Staphylococci Isolated from Hunan Smoked Pork, a Naturally Fermented Meat Product in China. J. Food Sci. 2018, 83, 1707–1715. [Google Scholar] [CrossRef]
- Nawaz, M.; Wang, J.; Zhou, A.; Ma, C.; Wu, X.; Moore, J.E.; Millar, B.C.; Xu, J. Characterization and transfer of antibiotic resistance in lactic acid bacteria from fermented food products. Curr. Microbiol. 2011, 62, 1081–1089. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Atienza, E.; Gómez-Sala, B.; Araújo, C.; Campanero, C.; del Campo, R.; Hernández, P.E.; Herranz, C.; Cintas, L.M. Antimicrobial activity, antibiotic susceptibility and virulence factors of Lactic Acid Bacteria of aquatic origin intended for use as probiotics in aquaculture. BMC Microbiol. 2013, 13, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, P.; Tomar, S.K.; Goswami, P.; Sangwan, V.; Singh, R. Antibiotic resistance among commercially available probiotics. Int. Food Res. J. 2014, 57, 176–195. [Google Scholar] [CrossRef]
- Kim, M.J.; Ku, S.; Kim, S.Y.; Lee, H.H.; Jin, H.; Kang, S.; Li, R.; Johnston, T.V.; Park, M.S.; Ji, G.E. Safety Evaluations of Bifidobacterium bifidum BGN4 and Bifidobacterium longum BORI. Int. J. Mol. Sci. 2018, 19, 1422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tenea, G.N.; Hurtado, P. Next-Generation Sequencing for Whole-Genome Characterization of Weissella cibaria UTNGt21O Strain Originated from Wild Solanum quitoense Lam. Fruits: An Atlas of Metabolites With Biotechnological Significance. Front. Microbiol. 2021, 12, 675002. [Google Scholar] [CrossRef]
- Monshupanee, T. Increased bacterial hemolytic activity is conferred by expression of TlyA methyltransferase but not by its 2′-O-methylation of the ribosome. Curr. Microbiol. 2013, 67, 61–68. [Google Scholar] [CrossRef]
- Beecher, D.J.; Schoeni, J.L.; Wong, A. Enterotoxic activity of hemolysin BL from Bacillus cereus. Infect. Immun. 1995, 63, 4423–4428. [Google Scholar] [CrossRef] [Green Version]
- Vitetta, L.; Coulson, S.; Thomsen, M.; Nguyen, T.; Hall, S. Probiotics, D-lactic acidosis, oxidative stress and strain specificity. Gut Microbes 2017, 8, 311–322. [Google Scholar] [CrossRef] [Green Version]
- Heavey, P.M.; Rowland, I.R. Microbial-gut interactions in health and disease. Gastrointestinal cancer. Best Pract. Res. Clin. Gastroenterol. 2004, 18, 323–336. [Google Scholar] [CrossRef]
- Ames, B.N.; Durston, W.E.; Yamasaki, E.; Lee, F.D. Carcinogens are mutagens: A simple test system combining liver homogenates for activation and bacteria for detection. Proc. Natl. Acad. Sci. USA 1973, 70, 2281. [Google Scholar] [CrossRef] [Green Version]
- Chiu, Y.J.; Nam, M.K.; Tsai, Y.T.; Huang, C.C.; Tsai, C.C. Genotoxicity assessment of multispecies probiotics using reverse mutation, mammalian chromosomal aberration, and rodent micronucleus tests. Sci. World J. 2013, 254239. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.C.; Chen, Y.T.; Chen, M.J. Lack of mutagenicity, genotoxicity and developmental toxicity in safety assessment tests of Lactobacillus mali APS1. PLoS ONE 2018, 13, e0208881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, H.; Zhao, W.; Liu, W.H.; Sun, T.; Lou, H.; Wei, T.; Hung, W.L.; Chen, Q. Safety Evaluation of Bifidobacterium lactis BL-99 and Lacticaseibacillus paracasei K56 and ET-22 in vitro and in vivo. Front. Microbiol. 2021, 12, 686541. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).