Identification of Novel Isatin Derivative Bearing a Nitrofuran Moiety as Potent Multi-Isoform Aldehyde Dehydrogenase Inhibitor
Abstract
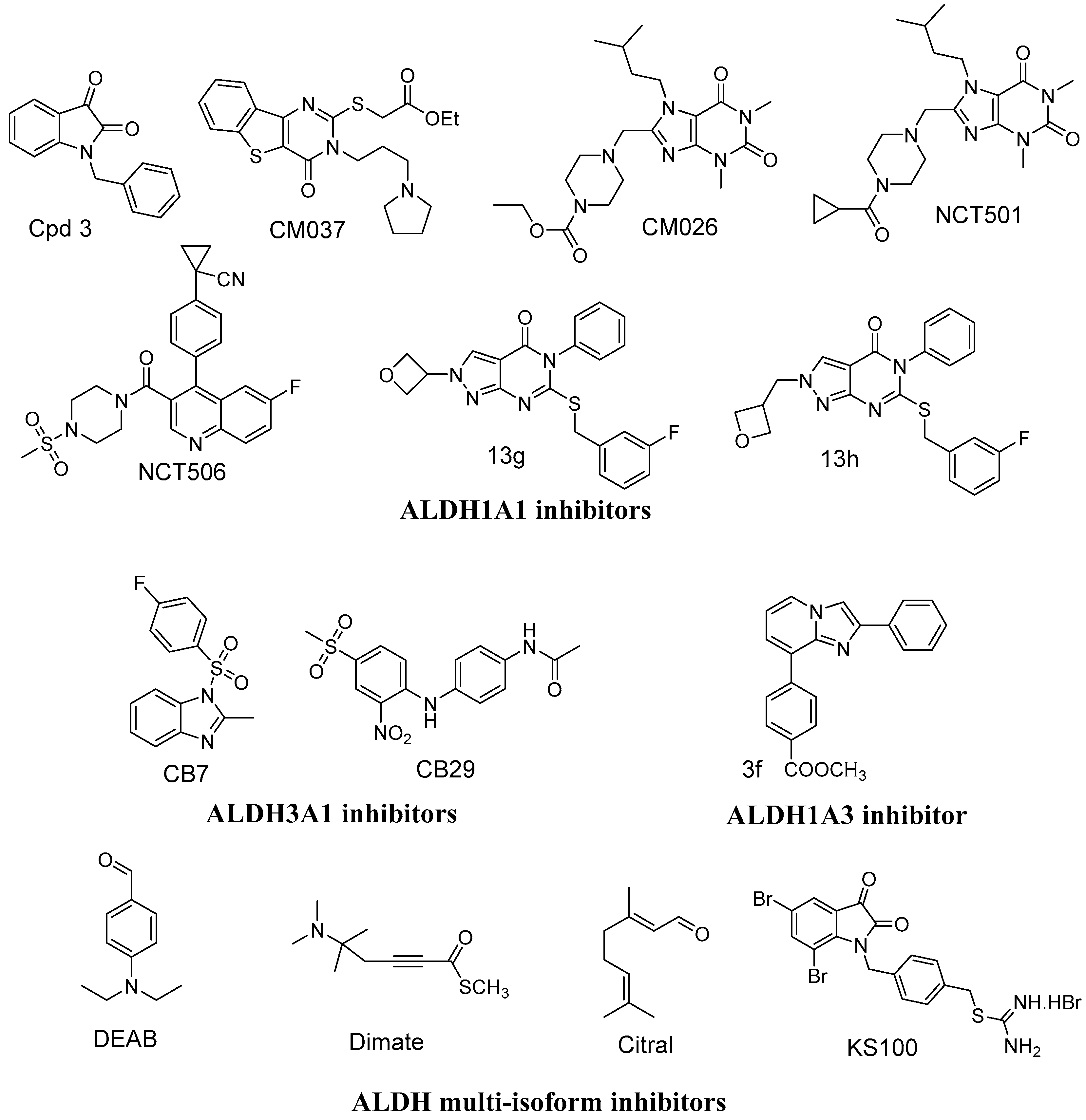
:1. Introduction
2. Results and Discussion
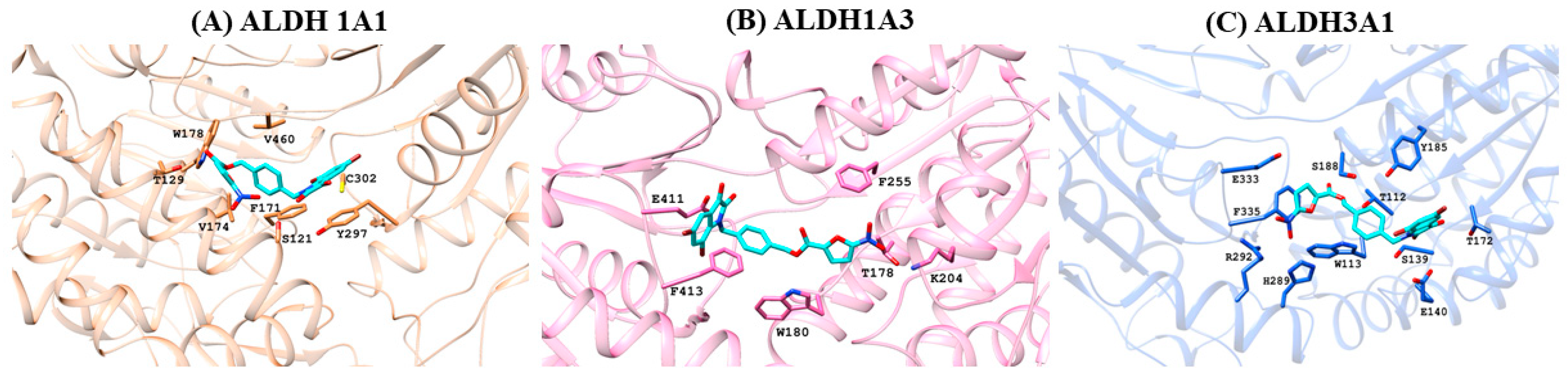
2.1. Molecular Docking Studies
2.2. Chemistry
2.3. Biological Evaluation
2.3.1. Cytotoxicity
2.3.2. ALDEFLUOR Assay and ALDH Isoform Inhibitory Activity
2.3.3. Evaluation of the Antiproliferative Activity of Compound 3 (MTS and TBDEA)
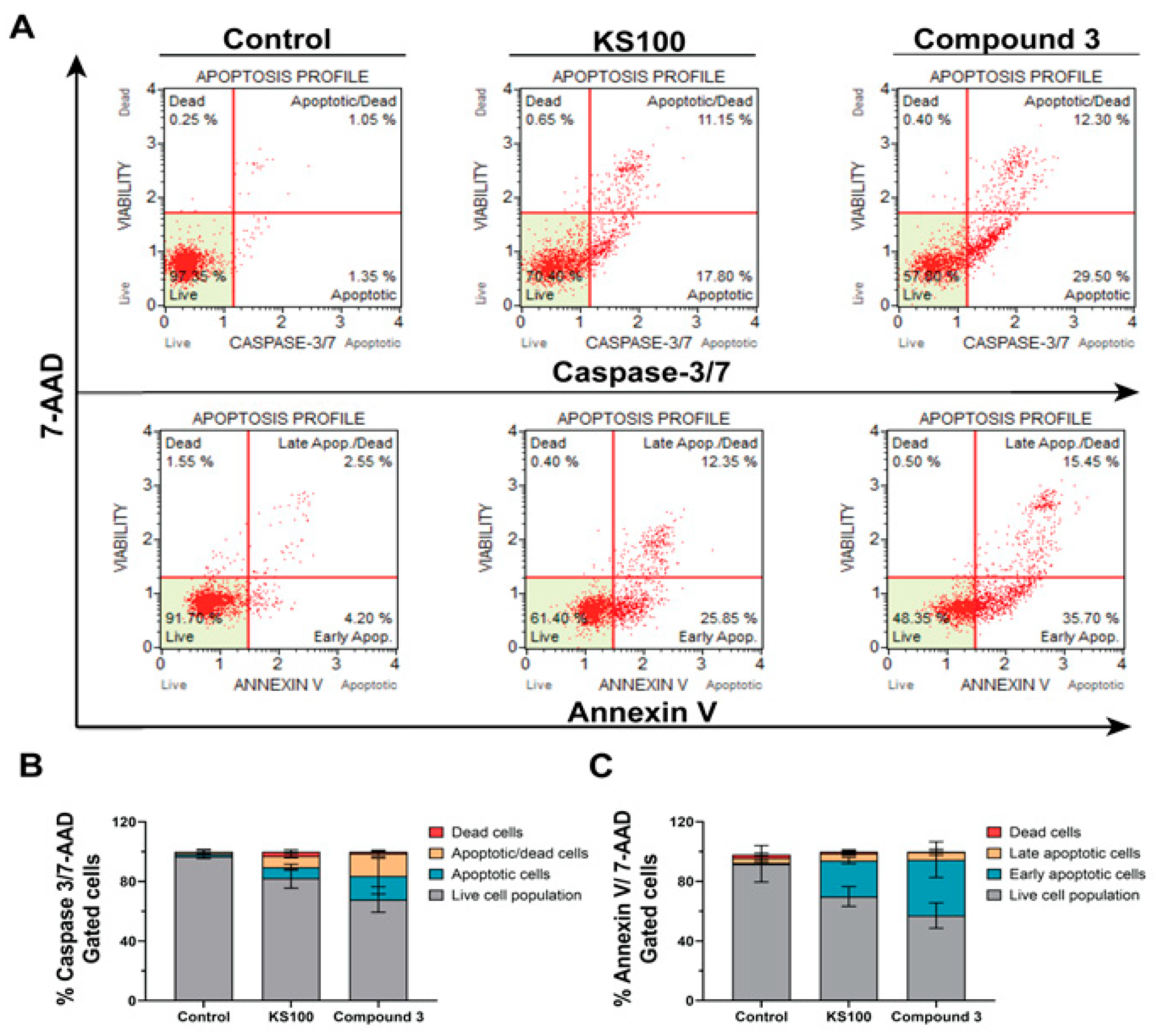
2.3.4. Compound 3 Induced Caspase-Mediated Apoptotic Cell Death in HCT116 Cells
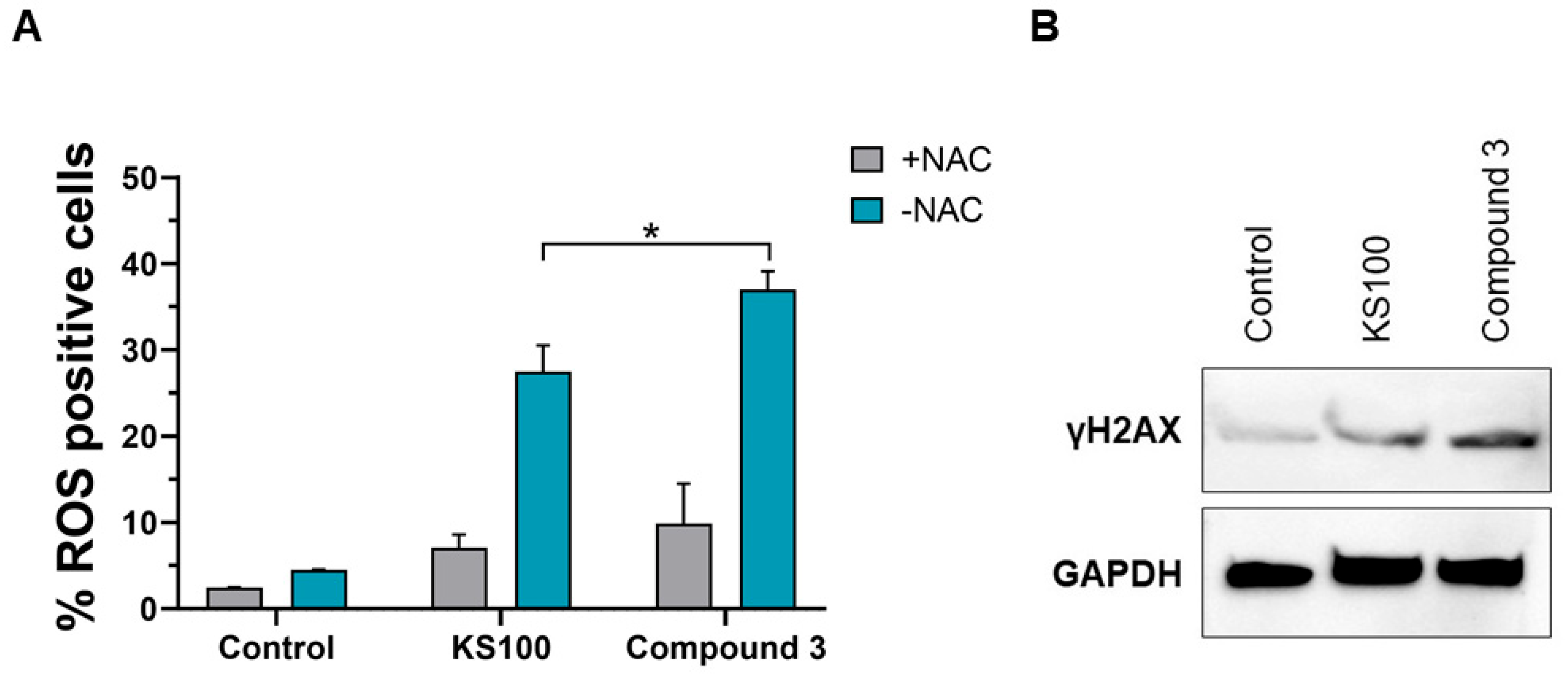
2.3.5. Compound 3 Generates more ROS Than KS100
3. Materials and Methods
3.1. Chemistry
Procedure for the Synthesis of Compounds (1–21)
3.2. Biological Evaluation
3.2.1. Cell Line and Culture Conditions
3.2.2. Molecular Docking Studies
3.2.3. ALDEFLUOR Assay
3.2.4. ALDH Isoform-Specific Enzyme Inhibition Assays
3.2.5. Cell Viability Assay
3.2.6. Trypan Blue Dye Exclusion Assay
3.2.7. Apoptosis Assay
3.2.8. Chemifluorescence Assay for Reactive Oxygen Species (ROS) Generation
3.2.9. Western Blot
3.2.10. Statistical Analysis
4. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Mizumoto, A.; Ohashi, S.; Hirohashi, K.; Amanuma, Y.; Matsuda, T.; Muto, M. Molecular Mechanisms of Acetaldehyde-Mediated Carcinogenesis in Squamous Epithelium. Int. J. Mol. Sci. 2017, 18, 1943. [Google Scholar] [CrossRef] [PubMed]
- Vasiliou, V.; Pappa, A.; Petersen, D.R. Role of aldehyde dehydrogenases in endogenous and xenobiotic metabolism. Chem. Biol. Interact. 2000, 129, 1–19. [Google Scholar] [CrossRef]
- O’Brien, P.J.; Siraki, A.G.; Shangari, N. Aldehyde sources, metabolism, molecular toxicity mechanisms, and possible effects on human health. Crit. Rev. Toxicol. 2005, 35, 609–662. [Google Scholar] [CrossRef]
- Esterbauer, H.; Schaur, R.J.; Zollner, H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic. Biol. Med. 1991, 11, 81–128. [Google Scholar] [CrossRef] [PubMed]
- Nadkarni, D.V.; Sayre, L.M. Structural definition of early lysine and histidine adduction chemistry of 4-hydroxynonenal. Chem. Res. Toxicol. 1995, 8, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Brooks, P.J.; Theruvathu, J.A. DNA adducts from acetaldehyde: Implications for alcohol-related carcinogenesis. Alcohol 2005, 35, 187–193. [Google Scholar] [CrossRef]
- LoPachin, R.M.; Gavin, T. Molecular mechanisms of aldehyde toxicity: A chemical perspective. Chem. Res. Toxicol. 2014, 27, 1081–1091. [Google Scholar] [CrossRef]
- Allison, S.E.; Chen, Y.; Petrovic, N.; Zhang, J.; Bourget, K.; Mackenzie, P.I.; Murray, M. Activation of ALDH1A1 in MDA-MB-468 breast cancer cells that over-express CYP2J2 protects against paclitaxel-dependent cell death mediated by reactive oxygen species. Biochem. Pharmacol. 2017, 143, 79–89. [Google Scholar] [CrossRef]
- Zanoni, M.; Bravaccini, S.; Fabbri, F.; Arienti, C. Emerging Roles of Aldehyde Dehydrogenase Isoforms in Anti-cancer Therapy Resistance. Front. Med. 2022, 9, 795762. [Google Scholar] [CrossRef]
- Ajani, J.A.; Wang, X.; Song, S.; Suzuki, A.; Taketa, T.; Sudo, K.; Wadhwa, R.; Hofstetter, W.L.; Komaki, R.; Maru, D.M.; et al. ALDH-1 expression levels predict response or resistance to preoperative chemoradiation in resectable esophageal cancer patients. Mol. Oncol. 2014, 8, 142–149. [Google Scholar] [CrossRef]
- Huang, C.P.; Tsai, M.F.; Chang, T.H.; Tang, W.C.; Chen, S.Y.; Lai, H.H.; Lin, T.Y.; Yang, J.C.; Yang, P.C.; Shih, J.Y.; et al. ALDH-positive lung cancer stem cells confer resistance to epidermal growth factor receptor tyrosine kinase inhibitors. Cancer Lett. 2013, 328, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Charafe-Jauffret, E.; Ginestier, C.; Iovino, F.; Tarpin, C.; Diebel, M.; Esterni, B.; Houvenaeghel, G.; Extra, J.M.; Bertucci, F.; Jacquemier, J.; et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin. Cancer Res. 2010, 16, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Marcato, P.; Dean, C.A.; Giacomantonio, C.A.; Lee, P.W. Aldehyde dehydrogenase: Its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle 2011, 10, 1378–1384. [Google Scholar] [CrossRef] [PubMed]
- Durinikova, E.; Kozovska, Z.; Poturnajova, M.; Plava, J.; Cierna, Z.; Babelova, A.; Bohovic, R.; Schmidtova, S.; Tomas, M.; Kucerova, L.; et al. ALDH1A3 upregulation and spontaneous metastasis formation is associated with acquired chemoresistance in colorectal cancer cells. BMC Cancer 2018, 18, 848. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, Q.; Liu, X.; Liu, C.; Liu, R.; Rycaj, K.; Zhang, D.; Liu, B.; Jeter, C.; Calhoun-Davis, T.; et al. Defining a Population of Stem-like Human Prostate Cancer Cells That Can Generate and Propagate Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2016, 22, 4505–4516. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Jou, D.; Wang, Y.; Ma, H.; Liu, T.; Fuchs, J.; Li, P.K.; Lu, J.; Li, C.; Lin, J. STAT3 as a potential therapeutic target in ALDH+ and CD44+/CD24+ stem cell-like pancreatic cancer cells. Int. J. Oncol. 2016, 49, 2265–2274. [Google Scholar] [CrossRef] [PubMed]
- Sarvi, S.; Crispin, R.; Lu, Y.; Zeng, L.; Hurley, T.D.; Houston, D.R.; von Kriegsheim, A.; Chen, C.H.; Mochly-Rosen, D.; Ranzani, M.; et al. ALDH1 Bio-activates Nifuroxazide to Eradicate ALDH(High) Melanoma-Initiating Cells. Cell Chem. Biol. 2018, 25, 1456–1469.e6. [Google Scholar] [CrossRef] [PubMed]
- Flahaut, M.; Jauquier, N.; Chevalier, N.; Nardou, K.; Balmas Bourloud, K.; Joseph, J.M.; Barras, D.; Widmann, C.; Gross, N.; Renella, R.; et al. Aldehyde dehydrogenase activity plays a Key role in the aggressive phenotype of neuroblastoma. BMC Cancer 2016, 16, 781. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Li, S.; Liu, S.; Zhang, L. Aldehyde dehydrogenase in solid tumors and other diseases: Potential biomarkers and therapeutic targets. MedComm 2023, 4, e195. [Google Scholar] [CrossRef]
- Muzio, G.; Maggiora, M.; Paiuzzi, E.; Oraldi, M.; Canuto, R.A. Aldehyde dehydrogenases and cell proliferation. Free Radic. Biol. Med. 2012, 52, 735–746. [Google Scholar] [CrossRef]
- Vlashi, E.; Pajonk, F. Cancer stem cells, cancer cell plasticity and radiation therapy. Semin. Cancer Biol. 2015, 31, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Bazewicz, C.G.; Dinavahi, S.S.; Schell, T.D.; Robertson, G.P. Aldehyde dehydrogenase in regulatory T-cell development, immunity and cancer. Immunology 2019, 156, 47–55. [Google Scholar] [CrossRef]
- Rodriguez-Zavala, J.S.; Calleja, L.F.; Moreno-Sanchez, R.; Yoval-Sanchez, B. Role of Aldehyde Dehydrogenases in Physiopathological Processes. Chem. Res. Toxicol. 2019, 32, 405–420. [Google Scholar] [CrossRef] [PubMed]
- Oh, T.; Kwon, M.; Yu, J.S.; Jang, M.; Kim, G.H.; Kim, K.H.; Ko, S.K.; Ahn, J.S. Ent-Peniciherqueinone Suppresses Acetaldehyde-Induced Cytotoxicity and Oxidative Stress by Inducing ALDH and Suppressing MAPK Signaling. Pharmaceutics 2020, 12, 1229. [Google Scholar] [CrossRef]
- McLean, M.E.; MacLean, M.R.; Cahill, H.F.; Arun, R.P.; Walker, O.L.; Wasson, M.D.; Fernando, W.; Venkatesh, J.; Marcato, P. The Expanding Role of Cancer Stem Cell Marker ALDH1A3 in Cancer and Beyond. Cancers 2023, 15, 492. [Google Scholar] [CrossRef]
- Koppaka, V.; Thompson, D.C.; Chen, Y.; Ellermann, M.; Nicolaou, K.C.; Juvonen, R.O.; Petersen, D.; Deitrich, R.A.; Hurley, T.D.; Vasiliou, V. Aldehyde dehydrogenase inhibitors: A comprehensive review of the pharmacology, mechanism of action, substrate specificity, and clinical application. Pharmacol. Rev. 2012, 64, 520–539. [Google Scholar] [CrossRef] [PubMed]
- Kimble-Hill, A.C.; Parajuli, B.; Chen, C.H.; Mochly-Rosen, D.; Hurley, T.D. Development of selective inhibitors for aldehyde dehydrogenases based on substituted indole-2,3-diones. J. Med. Chem. 2014, 57, 714–722. [Google Scholar] [CrossRef]
- Morgan, C.A.; Hurley, T.D. Characterization of two distinct structural classes of selective aldehyde dehydrogenase 1A1 inhibitors. J. Med. Chem. 2015, 58, 1964–1975. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.M.; Yasgar, A.; Miller, B.; Lal-Nag, M.; Brimacombe, K.; Hu, X.; Sun, H.; Wang, A.; Xu, X.; Nguyen, K.; et al. Discovery of NCT-501, a Potent and Selective Theophylline-Based Inhibitor of Aldehyde Dehydrogenase 1A1 (ALDH1A1). J. Med. Chem. 2015, 58, 5967–5978. [Google Scholar] [CrossRef]
- Yang, S.M.; Martinez, N.J.; Yasgar, A.; Danchik, C.; Johansson, C.; Wang, Y.; Baljinnyam, B.; Wang, A.Q.; Xu, X.; Shah, P.; et al. Discovery of Orally Bioavailable, Quinoline-Based Aldehyde Dehydrogenase 1A1 (ALDH1A1) Inhibitors with Potent Cellular Activity. J. Med. Chem. 2018, 61, 4883–4903. [Google Scholar] [CrossRef]
- Kulsum, S.; Sudheendra, H.V.; Pandian, R.; Ravindra, D.R.; Siddappa, G.; Chevour, P.; Ramachandran, B.; Sagar, M.; Jayaprakash, A.; Mehta, A.; et al. Cancer stem cell mediated acquired chemoresistance in head and neck cancer can be abrogated by aldehyde dehydrogenase 1 A1 inhibition. Mol. Carcinog. 2017, 56, 694–711. [Google Scholar] [CrossRef] [PubMed]
- Huddle, B.C.; Grimley, E.; Buchman, C.D.; Chtcherbinine, M.; Debnath, B.; Mehta, P.; Yang, K.; Morgan, C.A.; Li, S.; Felton, J.; et al. Structure-Based Optimization of a Novel Class of Aldehyde Dehydrogenase 1A (ALDH1A) Subfamily-Selective Inhibitors as Potential Adjuncts to Ovarian Cancer Chemotherapy. J. Med. Chem. 2018, 61, 8754–8773. [Google Scholar] [CrossRef] [PubMed]
- Parajuli, B.; Fishel, M.L.; Hurley, T.D. Selective ALDH3A1 inhibition by benzimidazole analogues increase mafosfamide sensitivity in cancer cells. J. Med. Chem. 2014, 57, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Yang, K.; Liang, D.; Jiang, C.; Ma, Z. Discovery and development of selective aldehyde dehydrogenase 1A1 (ALDH1A1) inhibitors. Eur. J. Med. Chem. 2021, 209, 112940. [Google Scholar] [CrossRef] [PubMed]
- Quattrini, L.; Gelardi, E.L.M.; Petrarolo, G.; Colombo, G.; Ferraris, D.M.; Picarazzi, F.; Rizzi, M.; Garavaglia, S.; La Motta, C. Progress in the Field of Aldehyde Dehydrogenase Inhibitors: Novel Imidazo [1,2-a]pyridines against the 1A Family. ACS Med. Chem. Lett. 2020, 11, 963–970. [Google Scholar] [CrossRef] [PubMed]
- Quattrini, L.; Gelardi, E.L.M.; Coviello, V.; Sartini, S.; Ferraris, D.M.; Mori, M.; Nakano, I.; Garavaglia, S.; La Motta, C. Imidazo [1,2-a]pyridine Derivatives as Aldehyde Dehydrogenase Inhibitors: Novel Chemotypes to Target Glioblastoma Stem Cells. J. Med. Chem. 2020, 63, 4603–4616. [Google Scholar] [CrossRef] [PubMed]
- Gelardi, E.L.M.; Colombo, G.; Picarazzi, F.; Ferraris, D.M.; Mangione, A.; Petrarolo, G.; Aronica, E.; Rizzi, M.; Mori, M.; La Motta, C.; et al. A Selective Competitive Inhibitor of Aldehyde Dehydrogenase 1A3 Hinders Cancer Cell Growth, Invasiveness and Stemness In Vitro. Cancers 2021, 13, 356. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, N.; Ogino, T.; Hara, Y.; Tanaka, T.; Koyanagi, S.; Ohdo, S. Optimized Dosing Schedule Based on Circadian Dynamics of Mouse Breast Cancer Stem Cells Improves the Antitumor Effects of Aldehyde Dehydrogenase Inhibitor. Cancer Res. 2018, 78, 3698–3708. [Google Scholar] [CrossRef] [PubMed]
- Morgan, C.A.; Parajuli, B.; Buchman, C.D.; Dria, K.; Hurley, T.D. N,N-diethylaminobenzaldehyde (DEAB) as a substrate and mechanism-based inhibitor for human ALDH isoenzymes. Chem. Biol. Interact. 2015, 234, 18–28. [Google Scholar] [CrossRef]
- Fournet, G.; Martin, G.; Quash, G. alpha,beta-Acetylenic amino thiolester inhibitors of aldehyde dehydrogenases 1&3: Suppressors of apoptogenic aldehyde oxidation and activators of apoptosis. Curr. Med. Chem. 2013, 20, 527–533. [Google Scholar] [CrossRef]
- Perez-Alea, M.; McGrail, K.; Sanchez-Redondo, S.; Ferrer, B.; Fournet, G.; Cortes, J.; Munoz, E.; Hernandez-Losa, J.; Tenbaum, S.; Martin, G.; et al. ALDH1A3 is epigenetically regulated during melanocyte transformation and is a target for melanoma treatment. Oncogene 2017, 36, 5695–5708. [Google Scholar] [CrossRef] [PubMed]
- Venton, G.; Perez-Alea, M.; Baier, C.; Fournet, G.; Quash, G.; Labiad, Y.; Martin, G.; Sanderson, F.; Poullin, P.; Suchon, P.; et al. Aldehyde dehydrogenases inhibition eradicates leukemia stem cells while sparing normal progenitors. Blood Cancer J. 2016, 6, e469. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.L.; de Antueno, R.; Coyle, K.M.; Sultan, M.; Cruickshank, B.M.; Giacomantonio, M.A.; Giacomantonio, C.A.; Duncan, R.; Marcato, P. Citral reduces breast tumor growth by inhibiting the cancer stem cell marker ALDH1A3. Mol. Oncol. 2016, 10, 1485–1496. [Google Scholar] [CrossRef] [PubMed]
- Khanna, M.; Chen, C.H.; Kimble-Hill, A.; Parajuli, B.; Perez-Miller, S.; Baskaran, S.; Kim, J.; Dria, K.; Vasiliou, V.; Mochly-Rosen, D.; et al. Discovery of a novel class of covalent inhibitor for aldehyde dehydrogenases. J. Biol. Chem. 2011, 286, 43486–43494. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Shin, J.H.; Chen, C.H.; Cruz, L.; Farnebo, L.; Yang, J.; Borges, P.; Kang, G.; Mochly-Rosen, D.; Sunwoo, J.B. Targeting aldehyde dehydrogenase activity in head and neck squamous cell carcinoma with a novel small molecule inhibitor. Oncotarget 2017, 8, 52345–52356. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, S.; Shintani, S.; Hirata, Y.; Suina, K.; Semba, T.; Yamasaki, J.; Umene, K.; Ishikawa, M.; Saya, H.; Nagano, O. Synthetic lethality of the ALDH3A1 inhibitor dyclonine and xCT inhibitors in glutathione deficiency-resistant cancer cells. Oncotarget 2018, 9, 33832–33843. [Google Scholar] [CrossRef]
- Zeng, S.; Kapur, A.; Patankar, M.S.; Xiong, M.P. Formulation, Characterization, and Antitumor Properties of Trans- and Cis-Citral in the 4T1 Breast Cancer Xenograft Mouse Model. Pharm. Res. 2015, 32, 2548–2558. [Google Scholar] [CrossRef] [PubMed]
- Dinavahi, S.S.; Gowda, R.; Bazewicz, C.G.; Battu, M.B.; Lin, J.M.; Chitren, R.J.; Pandey, M.K.; Amin, S.; Robertson, G.P.; Gowda, K. Design, synthesis characterization and biological evaluation of novel multi-isoform ALDH inhibitors as potential anticancer agents. Eur. J. Med. Chem. 2020, 187, 111962. [Google Scholar] [CrossRef] [PubMed]
- Dinavahi, S.S.; Gowda, R.; Gowda, K.; Bazewicz, C.G.; Chirasani, V.R.; Battu, M.B.; Berg, A.; Dokholyan, N.V.; Amin, S.; Robertson, G.P. Development of a Novel Multi-Isoform ALDH Inhibitor Effective as an Antimelanoma Agent. Mol. Cancer Ther. 2020, 19, 447–459. [Google Scholar] [CrossRef]
- Jackson, B.; Brocker, C.; Thompson, D.C.; Black, W.; Vasiliou, K.; Nebert, D.W.; Vasiliou, V. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum. Genom. 2011, 5, 283–303. [Google Scholar] [CrossRef]
- Januchowski, R.; Wojtowicz, K.; Zabel, M. The role of aldehyde dehydrogenase (ALDH) in cancer drug resistance. Biomed. Pharmacother. 2013, 67, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Dinavahi, S.S.; Bazewicz, C.G.; Gowda, R.; Robertson, G.P. Aldehyde Dehydrogenase Inhibitors for Cancer Therapeutics. Trends Pharmacol. Sci. 2019, 40, 774–789. [Google Scholar] [CrossRef] [PubMed]
- Chefetz, I.; Grimley, E.; Yang, K.; Hong, L.; Vinogradova, E.V.; Suciu, R.; Kovalenko, I.; Karnak, D.; Morgan, C.A.; Chtcherbinine, M.; et al. A Pan-ALDH1A Inhibitor Induces Necroptosis in Ovarian Cancer Stem-like Cells. Cell Rep. 2019, 26, 3061–3075.e6. [Google Scholar] [CrossRef] [PubMed]
- Van Engeland, M.; Nieland, L.J.; Ramaekers, F.C.; Schutte, B.; Reutelingsperger, C.P. Annexin V-affinity assay: A review on an apoptosis detection system based on phosphatidylserine exposure. Cytom. J. Int. Soc. Anal. Cytol. 1998, 31, 1–9. [Google Scholar] [CrossRef]
- Ozben, T. Oxidative stress and apoptosis: Impact on cancer therapy. J. Pharm. Sci. 2007, 96, 2181–2196. [Google Scholar] [CrossRef] [PubMed]
- Zhitkovich, A. N-Acetylcysteine: Antioxidant, Aldehyde Scavenger, and More. Chem. Res. Toxicol. 2019, 32, 1318–1319. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Nikhil, K.; Raza, A.; Haymour, H.S.; Flueckiger, B.V.; Chu, J.; Shah, K. Aurora Kinase A-YBX1 Synergy Fuels Aggressive Oncogenic Phenotypes and Chemoresistance in Castration-Resistant Prostate Cancer. Cancers 2020, 12, 660. [Google Scholar] [CrossRef]




| Docking Score (kcal/mol) | |||
|---|---|---|---|
| Compound | ALDH1A1 | ALDH1A3 | ALDH3A1 |
| 1 | −8.5 | −8.3 | −7.2 |
| 2 | −8.8 | −8.5 | −8.3 |
| 3 | −10.1 | −9.6 | −10.4 |
| 4 | −9.2 | −7.9 | −7.9 |
| 5 | −8.2 | −7.4 | −7.8 |
| 6 | −8.7 | −8.1 | −7.7 |
| 7 | −9.5 | −8.2 | −8.5 |
| 8 | −9.2 | −8.6 | −8.7 |
| 9 | −9.2 | −8.4 | −8.5 |
| 10 | −9.2 | −8.2 | −8.6 |
| 11 | −8.9 | −8.2 | −8.7 |
| 12 | −8.9 | −8.2 | −9.0 |
| 13 | −9.4 | −7.2 | −7.5 |
| 14 | −8.0 | −7.2 | −8.3 |
| 15 | −9.9 | −8.5 | −9.0 |
| 16 | −9.2 | −8.1 | −8.1 |
| 17 | −8.2 | −7.9 | −7.7 |
| 18 | −9.5 | −8.8 | −8.2 |
| 19 | −10.1 | −9.1 | −8.5 |
| 20 | −10.0 | −9.5 | −9.1 |
| 21 | −9.7 | −8.6 | −9.8 |
| KS100 | −7.9 | −7.4 | −8.3 |
| CM037 | −8.0 | −7.6 | −7.9 |
 Compound 8 Compound 8 | 1H NMR (500 MHz, DMSO-d6) δ: H1 = 8.33 (s, 1H), H4 = 7.81 (d, J = 8.95 Hz, 1H), H5 = 7.45 (m, 1H), H7 = 8.27 (s, 1H), H10 = 5.79 (s, 2H), H12 = 7.26 (d, J = 8.23 Hz, 2H), H13 = 7.61 (d, J = 8.23 Hz, 2H), H15 = 7.61 (d, J = 16.13 Hz, 1H), H16 = 6.60 (d, J = 16.13 Hz, 1H), H18 = 3.71 (s, 3H). |
| 13C NMR (125 MHz, DMSO-d6) δ: C17 = 167.05, C15 = 144.41, C9 = 140.82, C11 = 139.9, C1 = 135.34, C14 = 133.94, C13 = 129.10, CF3 = 125.36 (q, J = 271.44 Hz), C8 = 123.42, C7 = 123.08 (q, J = 2.95 Hz), C6 = 122.01 (q, J = 31.54 Hz), C5 = 120.06 (q, J = 4.60 Hz), C12 = 128.30, C16 = 118.42, C4 = 111.56, C10 = 52.17, C18 = 51.93 | |
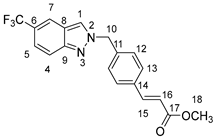 Compound 9 Compound 9 | 1H NMR (500 MHz, DMSO-d6) δ: H1 = 8.77 (s, 1H), H7 = 8.26 (s, 1H), H4 = 7.81 (d, J = 8.91 Hz, 1H), H13 = 7.72 (d, J = 8.23 Hz, 2H), H15 = 7.64 (d, J = 16.10 Hz, 1H), H5 = 7.45 (dd, J = 8.91 Hz, J = 1.59 Hz, 1H), H12 = 7.37 (d, J = 8.23 Hz, 2H), H16 = 6.63 (d, J = 16.10 Hz, 1H), H10 = 5.76 (s, 2H), H18 = 3.72 (s, 3H). |
| 13C NMR (125 MHz, DMSO-d6) δ: C17 = 167.06, C9 = 149.13, C4 = 144.37, C11 = 139.27, C14 = 134.23, C13 = 129.11, C12 = 128.77, C1 = 127.47, CF3 = 125.41 (q, J = 271.62 Hz), C6 = 122.20 (q, J = 31.37 Hz), C5 = 121.67 (q, J = 2.93 Hz), C7 = 120.68 (q, J = 5.05 Hz), C8 = 120.59, C15 = 118.99, C16 = 118.64, C10 = 56.65, C18 = 51.95 |
| Compound No. | Structure | a IC50 (µM) | ||||
|---|---|---|---|---|---|---|
| b SKOV-3 | c MiaPaCa-2 | d HCT-116 | e HEK-293 | f FHC | ||
| 1 | 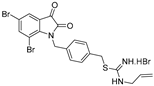 | 4.8 ± 0.2 | 2.4 ± 1.1 | 4.7 ± 1.1 | 4.1 ± 0.8 | 2.9 ± 0.3 |
| 2 | 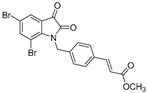 | 21.8 ± 2.4 | 22.3 ± 5.1 | 19.6 ± 3.3 | 24.1 ± 6.8 | 22.9 ± 3.3 |
| 3 (KS124) | 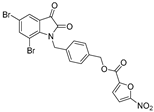 | 2.2 ± 0.2 | 3.8 ± 0.4 | 2.1 ± 0.1 | 8.7 ± 1.1 | 7.1 ± 0.8 |
| 4 |  | 7.2 ± 1.3 | 7.1 ± 0.8 | 6.1 ± 1.2 | 5.3 ± 1.4 | 6.7 ± 1.9 |
| 5 |  | 8.2 ± 1.1 | 5.4 ± 0.9 | 8.8 ± 2.3 | 7.5 ± 1.8 | 12.9 ± 3.3 |
| 6 | 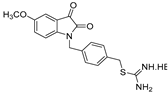 | 45.0 ± 2.5 | >50 | >50 | >50 | >50 |
| 7 | 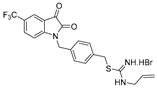 | 9.1 ± 1.3 | 10.3 ± 2.4 | 12.0 ± 3.1 | 9.7 ± 1.1 | 8.1 ± 0.8 |
| 8 | 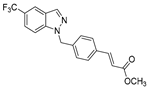 | >50 | >50 | >50 | >50 | >50 |
| 9 | 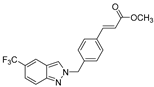 | >50 | >50 | >50 | >50 | >50 |
| 10 |  | >50 | 48.4 ± 3.2 | >50 | >50 | >50 |
| 11 | 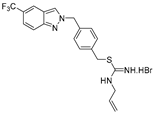 | >50 | >50 | >50 | >50 | >50 |
| 12 | 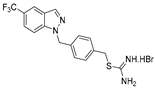 | 8.2 ± 2.7 | 4.7 ± 0.4 | 5.5 ± 0.1 | 4.7 ± 1.1 | 3.1 ± 0.8 |
| 13 | 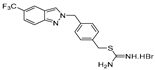 | 22.7 ± 2.3 | 33.2 ± 4.4 | 18.2 ± 3.1 | 18.9 ± 5.1 | 14.4 ± 2.3 |
| 14 | 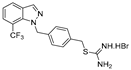 | 16.8 ± 3.5 | 12.3 ± 2.7 | 14.2 ± 2.3 | 17.7 ± 3.7 | 29.3 ± 4.9 |
| 15 | 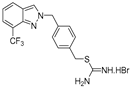 | >50 | >50 | >50 | >50 | >50 |
| 16 |  | 4.0 ± 1.3 | 6.8 ± 1.4 | 4.9 ± 1.1 | 5.7 ± 1.3 | 7.1 ± 1.1 |
| 17 | 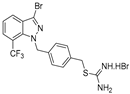 | >50 | >50 | >50 | >50 | >50 |
| 18 | 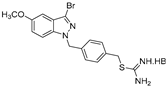 | 42.2 ± 5.8 | 44.6 ± 3.7 | 39.2 ± 3.9 | >50 | >50 |
| 19 | 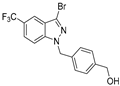 | >50 | >50 | >50 | >50 | >50 |
| 20 | 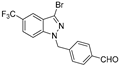 | >50 | >50 | >50 | >50 | >50 |
| 21 | 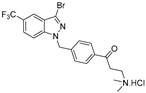 | >50 | >50 | >50 | >50 | >50 |
| KS100 | 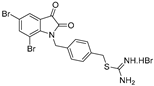 | 3.8 ± 0.8 | 2.8 ± 0.2 | 4.3 ± 0.7 | 4.3 ± 0.1 | 4.9 ± 0.8 |
| Compounds | % Enzyme Inhibition at 500 nM | ||
|---|---|---|---|
| Acetaldehyde (Substrate) | 4-Nitrobenzaldeyde (Substrate) | ||
| ALDH1A1 | ALDH3A1 | ALDH1A3 | |
| Control | 0 ± 0.02 | 0.05 ± 0.05 | 0 ± 0.01 |
| 1 | 35.37 ± 0.01 | 30.59 ± 0.02 | ND |
| 3 (KS124) | 51.32 ± 0.01 | 51.87 ± 0.05 | 36.65 ± 0.02 |
| 4 | 28.26 ± 0.01 | 22.82 ± 0.05 | 35.14 ± 0.07 |
| 5 | 24.82 ± 0.01 | 39.58 ± 0.05 | ND |
| 12 | 10.01 ± 0.01 | −14.03 ± 0.06 | ND |
| 16 | 9.46 ± 0.01 | −9.33 ± 0.04 | ND |
| 17 | 10.99 ± 0.0015 | −25.33 ± 0.05 | ND |
| KS100 | 45.93 ± 0.01 | 37.04 ± 0.09 | 13.83 ± 0.01 |
| 673A-IC50 [53] | 246 nM | ND | 348 nM |
| CB7-IC50 [33] | ND | 200 nM | ND |
| Configuration parameter | |||
|---|---|---|---|
| Config | ALDH1A1 (4X4L) | ALDH1A3 (7QK7) | ALDH3A1 (3SZB) |
| center_x | −25.6 | 22.0 | 7.9 |
| center_y | 38.1 | 11.5 | 17.7 |
| center_z | 30.2 | 23.9 | 66.7 |
| size_x | 40 | 40 | 40 |
| size_y | 40 | 40 | 40 |
| size_z | 40 | 40 | 40 |
| exhaustiveness | 32 | 32 | 32 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gowda, K.; Raza, A.; Vangala, V.; Lone, N.A.; Lin, J.M.; Singh, J.K.; Srivastava, S.K.; Schell, T.D.; Robertson, G.P.; Amin, S.; et al. Identification of Novel Isatin Derivative Bearing a Nitrofuran Moiety as Potent Multi-Isoform Aldehyde Dehydrogenase Inhibitor. Molecules 2024, 29, 3114. https://doi.org/10.3390/molecules29133114
Gowda K, Raza A, Vangala V, Lone NA, Lin JM, Singh JK, Srivastava SK, Schell TD, Robertson GP, Amin S, et al. Identification of Novel Isatin Derivative Bearing a Nitrofuran Moiety as Potent Multi-Isoform Aldehyde Dehydrogenase Inhibitor. Molecules. 2024; 29(13):3114. https://doi.org/10.3390/molecules29133114
Chicago/Turabian StyleGowda, Krishne, Asif Raza, Venugopal Vangala, Nazir Ahmad Lone, Jyh Ming Lin, Jaikee Kumar Singh, Sandeep Kumar Srivastava, Todd D. Schell, Gavin P. Robertson, Shantu Amin, and et al. 2024. "Identification of Novel Isatin Derivative Bearing a Nitrofuran Moiety as Potent Multi-Isoform Aldehyde Dehydrogenase Inhibitor" Molecules 29, no. 13: 3114. https://doi.org/10.3390/molecules29133114
APA StyleGowda, K., Raza, A., Vangala, V., Lone, N. A., Lin, J. M., Singh, J. K., Srivastava, S. K., Schell, T. D., Robertson, G. P., Amin, S., & Sharma, A. K. (2024). Identification of Novel Isatin Derivative Bearing a Nitrofuran Moiety as Potent Multi-Isoform Aldehyde Dehydrogenase Inhibitor. Molecules, 29(13), 3114. https://doi.org/10.3390/molecules29133114








