Detection of Human Adenovirus and Rotavirus in Wastewater in Lusaka, Zambia: Demonstrating the Utility of Environmental Surveillance for the Community
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Sample Collection
2.3. Viral Concentration
2.3.1. Skimmed Milk Flocculation
2.3.2. Bag-Mediated Filtration System (BMFS)
Secondary Concentration Procedure
2.3.3. Polyethylene Glycol-Based (PEG) Concentration
2.4. Nucleic Acid Extraction
2.5. Genomic Screening for Adenovirus and Rotavirus
2.5.1. Detection of Adenovirus and Rotavirus on PCR
2.5.2. Conventional PCR and Sequencing of Adenovirus
2.6. Statistical Analysis
3. Results
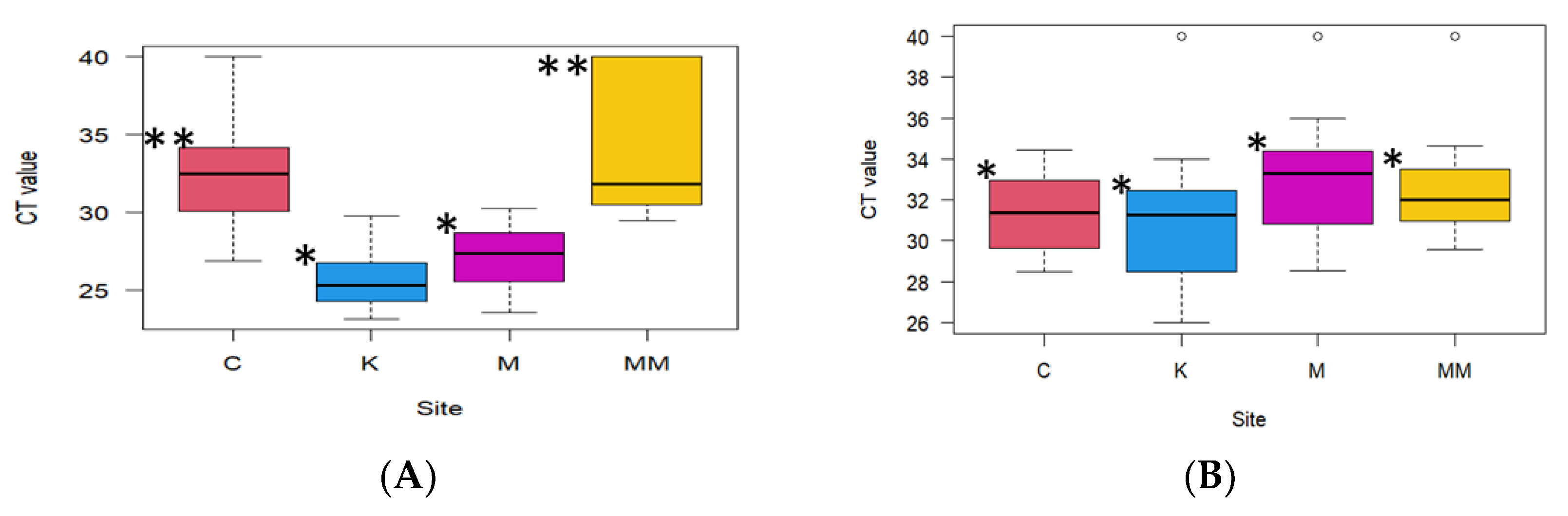
3.1. Detection of Adenovirus and Rotavirus
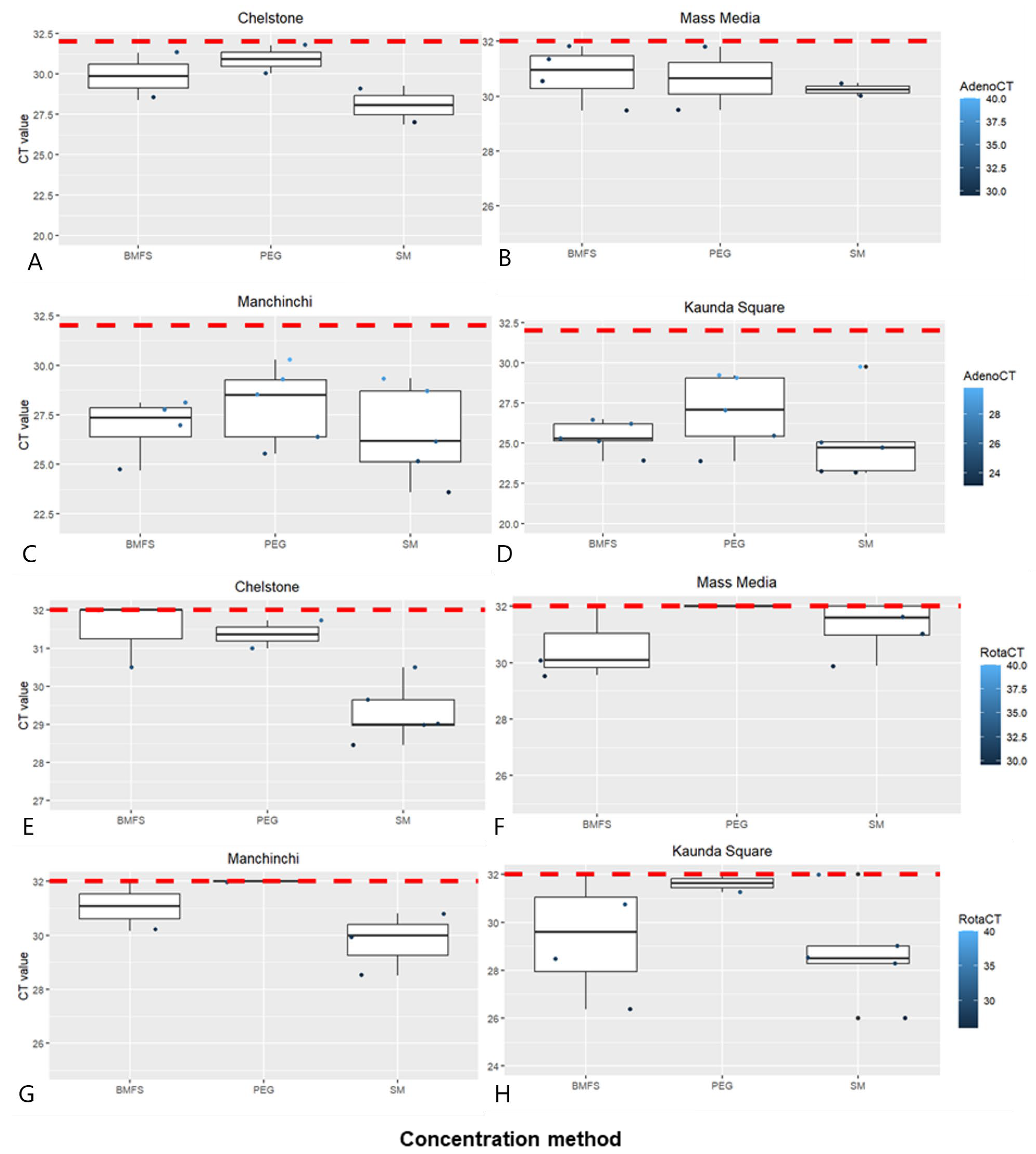
3.2. Comparison of Viral Concentration Methods
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fong, T.T.; Phanikumar, M.S.; Xagoraraki, I.; Rose, J.B. Quantitative detection of human adenoviruses in wastewater and combined sewer overflows influencing a Michigan river. Appl. Environ. Microbiol. 2010, 76, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Ali, W.; Zhang, H.; Wang, Z.; Chang, C.; Javed, A.; Ali, K.; Du, W.; Niazi, N.K.; Mao, K.; Yang, Z. Occurrence of various viruses and recent evidence of SARS-CoV-2 in wastewater systems. J. Hazard. Mater. 2021, 414, 125439. [Google Scholar] [CrossRef] [PubMed]
- Allayeh, A.K.; Al-Daim, S.A.; Ahmed, N.; El-Gayar, M.; Mostafa, A. Isolation and Genotyping of Adenoviruses from Wastewater and Diarrheal Samples in Egypt from 2016 to 2020. Viruses 2022, 14, 2192. [Google Scholar] [CrossRef] [PubMed]
- Osuolale, O.; Okoh, A. Incidence of human adenoviruses and Hepatitis A virus in the final effluent of selected wastewater treatment plants in Eastern Cape Province, South Africa. Virol. J. 2015, 12, 1–8. [Google Scholar] [CrossRef]
- El-Senousy, W.M.; Barakat, A.B.; Ghanem, H.E.; Kamel, M.A. Molecular epidemiology of human adenoviruses and rotaviruses as candidate viral indicators in the Egyptian sewage and water samples. World Appl. Sci. J. 2013, 27, 1235–1247. [Google Scholar] [CrossRef]
- Loevinsohn, G.; Hamahuwa, M.; Hardick, J.; Sinywimaanzi, P.; Fenstermacher, K.Z.J.; Munachoonga, P.; Weynand, A.; Monze, M.; Manabe, Y.C.; Gaydos, C.A.; et al. Respiratory viruses in rural Zambia before and during the COVID-19 pandemic. Trop. Med. Int. Health 2022, 27, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Soltane, R.; Allayeh, A.K. Occurrence of enteroviruses, noroviruses, rotaviruses, and adenoviruses in a wastewater treatment plant. J. Umm Al-Qura Univ. Appl. Sci. 2023, 9, 449–454. [Google Scholar] [CrossRef]
- Fletcher, S.M.; McLaws, M.-L.; Ellis, J.T. Prevalence of Gastrointestinal Pathogens in Developed and Developing Countries: Systematic Review and Meta-Analysis. J. Public Health Res. 2013, 2, jphr-2013. [Google Scholar] [CrossRef] [PubMed]
- Adefisoye, M.A.; Nwodo, U.U.; Green, E.; Okoh, A.I. Quantitative PCR Detection and Characterisation of Human Adenovirus, Rotavirus and Hepatitis A Virus in Discharged Effluents of Two Wastewater Treatment Facilities in the Eastern Cape, South Africa. Food Environ. Virol. 2016, 8, 262–274. [Google Scholar] [CrossRef] [PubMed]
- Mpabalwani, E.M.; Simwaka, C.J.; Mwenda, J.M.; Mubanga, C.P.; Monze, M.; Matapo, B.; Parashar, U.D.; Tate, J.E. Impact of Rotavirus Vaccination on Diarrheal Hospitalizations in Children Aged <5 Years in Lusaka, Zambia. Clin. Infect. Dis. 2016, 62, S183–S187. [Google Scholar] [CrossRef] [PubMed]
- Steele, A.; Victor, J.; Carey, M.; Tate, J.; Atherly, D.; Pecenka, C.; Diaz, Z.; Parashar, U.; Kirkwood, C. Experiences with rotavirus vaccines: Can we improve rotavirus vaccine impact in developing countries? Hum. Vaccin. Immunother. 2019, 15, 1215–1227. [Google Scholar] [CrossRef] [PubMed]
- Duncan Steele, A.; Groome, M.J. Measuring rotavirus vaccine impact in Sub-Saharan Africa. Clin. Infect. Dis. 2020, 70, 2314–2316. [Google Scholar] [CrossRef] [PubMed]
- Beres, L.K.; Tate, J.E.; Njobvu, L.; Chibwe, B.; Rudd, C.; Guffey, M.B.; Stringer, J.S.A.; Parashar, U.D.; Chilengi, R. A Preliminary Assessment of Rotavirus Vaccine Effectiveness in Zambia. Clin. Infect. Dis. 2016, 62, S175–S182. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, R.G.; Choi, C.Y.; Riley, M.R.; Gerba, C.P. Chapter 9 Pathogen Surveillance Through Monitoring of Sewer Systems. Adv. Appl. Microbiol. 2008, 65, 249. [Google Scholar] [CrossRef] [PubMed]
- Xagoraraki, I.; O’Brien, E. Wastewater-Based Epidemiology for Early Detection of Viral Outbreaks. In Women in Water Quality; O’Bannon, D.J., Ed.; Springer International Publishing: New York, NY, USA, 2020. [Google Scholar] [CrossRef]
- Qi, R.; Huang, Y.T.; Liu, J.W.; Sun, Y.; Sun, X.F.; Han, H.J.; Qin, X.R.; Zhao, M.; Wang, L.J.; Li, W.; et al. Global Prevalence of Asymptomatic Norovirus Infection: A Meta-analysis. EClinicalMedicine 2018, 2–3, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, M.; Ahmed, W.; Bibby, K.; Carducci, A.; Gerba, C.P.; Hamilton, K.A.; Haramoto, E.; Rose, J.B. SARS-CoV-2 in wastewater: State of the knowledge and research needs. Sci. Total Environ. 2020, 739, 139076. [Google Scholar] [CrossRef] [PubMed]
- Savolainen-Kopra, C.; Paananen, A.; Blomqvist, S.; Klemola, P.; Simonen, M.-L.; Lappalainen, M.; Vuorinen, T.; Kuusi, M.; Lemey, P.; Roivainen, M. A large Finnish echovirus 30 outbreak was preceded by silent circulation of the same genotype. Virus Genes 2011, 42, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Hellmér, M.; Paxéus, N.; Magnius, L.O.; Enache, L.; Arnholm, B.; Johansson, A.M.; Bergström, T.; Norder, H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014, 80, 6771–6781. [Google Scholar] [CrossRef] [PubMed]
- Prevost, B.; Lucas, F.S.; Goncalves, A.; Richard, F.; Moulin, L.; Wurtzer, S. Large scale survey of enteric viruses in river and waste water underlines the health status of the local population. Environ. Int. 2015, 79, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Shachakanza, J.; Zulu, J.M.; Maimbolwa, M. Incidence of Rotavirus Infection among Under-Five Children Attending Health Centres in Selected Communities of Ndola, Copperbelt Province, Zambia. Health N. Hav. 2019, 11, 298–307. [Google Scholar] [CrossRef]
- Chisenga, C.C.; Bosomprah, S.; Laban, N.M.; Kazimbaya, K.M.; Mwaba, J.; Simuyandi, M.; Chilengi, R. vAetiology of Diarrhoea in Children Under Five in Zambia Detected Using Luminex xTAG Gastrointestinal Pathogen Panel. Pediatr. Infect. Dis. Open Access 2018, 3, 1–6. [Google Scholar] [CrossRef]
- Prata, C.; Ribeiro, A.; Cunha, Â.; Gomes, N.C.M.; Almeida, A. Ultracentrifugation as a direct method to concentrate viruses in environmental waters: Virus-like particle enumeration as a new approach to determine the efficiency of recovery. J. Environ. Monit. 2012, 14, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Fagnant, C.S.; Sanchez-Gonzalez, L.M.; Zhou, N.A.; Falman, J.C.; Eisenstein, M.; Guelig, D.; Ockerman, B.; Guan, Y.; Kossik, A.L.; Linden, Y.S.; et al. Improvement of the Bag-Mediated Filtration System for Sampling Wastewater and Wastewater-Impacted Waters. Food Environ. Virol. 2018, 10, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Philo, S.E.; Keim, E.K.; Swanstrom, R.; Ong, A.Q.W.; Burnor, E.A.; Kossik, A.L.; Harrison, J.C.; Demeke, B.A.; Zhou, N.A.; Beck, N.K.; et al. A comparison of SARS-CoV-2 wastewater concentration methods for environmental surveillance. Sci. Total Environ. 2021, 760, 144215. [Google Scholar] [CrossRef]
- Jothikumar, N.; Cromeans, T.L.; Hill, V.R.; Lu, X.; Sobsey, M.D.; Erdman, D.D. Quantitative real-time PCR assays for detection of human adenoviruses and identification of serotypes 40 and 41. Appl. Environ. Microbiol. 2005, 71, 3131–3136. [Google Scholar] [CrossRef] [PubMed]
- Fumian, T.M.; Leite, J.P.G.; Castello, A.A.; Gaggero, A.; Caillou, M.S.L.; de Miagostovich, M.P. Detection of rotavirus A in sewage samples using multiplex qPCR and an evaluation of the ultracentrifugation and adsorption-elution methods for virus concentration. J. Virol. Methods 2010, 170, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Elmahdy, E.M.; Ahmed, N.I.; Shaheen, M.N.F.; Mohamed, E.C.B.; Loutfy, S.A. Molecular detection of human adenovirus in urban wastewater in Egypt and among children suffering from acute gastroenteritis. J. Water Health 2019, 17, 287–294. [Google Scholar] [CrossRef]
- Atabakhsh, P.; Kargar, M.; Doosti, A. Detection and evaluation of rotavirus surveillance methods as viral indicator in the aquatic environments. Braz. J. Microbiol. 2021, 52, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Haramoto, E.; Malla, B.; Thakali, O.; Kitajima, M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020, 737, 140405. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, G.B.; Mancini, P.; Veneri, C.; Iaconelli, M.; Suffredini, E.; Brandtner, D.; La Rosa, G. Evidence of Saffold virus circulation in Italy provided through environmental surveillance. Lett. Appl. Microbiol. 2020, 70, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Asghar, H.; Diop, O.M.; Weldegebriel, G.; Malik, F.; Shetty, S.; Bassioni, L.E.; Akande, A.O.; Maamoun, E.A.; Zaidi, S.; Adeniji, A.J.; et al. Environmental surveillance for polioviruses in the global polio eradication initiative. J. Infect. Dis. 2014, 210, S294–S303. [Google Scholar] [CrossRef]
| Site | Sewer Catchment Area |
|---|---|
| Chelstone pumping station | Chelstone Zambia Airways, Waterfalls mall |
| Kaunda Square stabilization ponds | PHI, Nyumba Yanga, Ibex Hills, Chamba Valley, Chainama, Kaunda Square Stage 1, Kaunda Square stage 2, Mtendere East, Salama Park, Mtendere, part of Kabulonga, part of Woodlands, Helen Kaunda, part of Kalingalinga, East Park mall, part of Handsworth |
| Mass Media pumping station | University of Zambia, part of Showgrounds, part of Longacres, Mass Media, Arcades mall up to National Assembly |
| Manchichi sewage treatment plant and Garden stabilization ponds | Part of Woodlands, Chilenje, Libala, Villa Elizabetha, Kabwata, Northmead, Parts of Emmasdale, part of Rhodes Park, Mass Media, part of Central Business district, Thorn Park, Part of the Light Industrial Area bordered by Great North Road, Sheki Road, Lumumba Road, and Kalambo Road. |
| No. | Name | Sequence | Tm (°C) | Reference |
|---|---|---|---|---|
| 1 | JTVXF_18895Fw | 5′-GGACGCCTCGGAGTACCTGAG-3′ | [26] | |
| 2 | JTVXR_18990Rv | 5′-ACIGTGGGGGTTTCTGAACTTGTT-3′ | 60 | |
| 3 | Probe JTVXP_18923Fw | 5′-CTGGTGCAGTTCGCCCGTGCCA-3′ | ||
| 4 | Hex1DegFw(1) | 5′-GCCSCARTGGKCWTACATGCACATC-3′ | 54 | [23] |
| 5 | Hex2DegRv(1) | 5′-CAGCACSCCICGRATGTCAAA-3′ | ||
| 6 | Adeno Cap 17467Fw(2) | 5′-CTGTAGGTTCCGTTCCCGTT-3′ | 54 | |
| 7 | Adeno Hex 18185Rv (2) | 5′-CGGTGCCTGAGCAAAGGTAT-3′ | ||
| 8 | RotaA-NSP3Fw | 5′-ACCATCTWCACRTRACCCTCTATGAG-3′ | [27] | |
| 9 | RotaA-NSP3Rv | 5′-GGTCACATAACGCCCCTATAGC-3′ | 60 | |
| 10 | Probe RotaA-NSP3Pr | 5′-AGTTAAAAGCTAACACTGTCAAA-3′ |
| Site | qPCR HAdV | cPCR HAdV | qPCR RVA |
|---|---|---|---|
| Chelstone | 3/5 | 3/5 | 3/5 |
| Mass Media | 5/5 | 3/5 | 4/5 |
| Kaunda Square | 5/5 | 5/5 | 4/5 |
| Manchichi | 5/5 | 5/5 | 3/5 |
| Total | 18/20 (90%) | 16/20 (80%) | 14/20 (70%) |
| Sample ID | Sampling Site | Week of Collection | RVA qPCR | HAdV qPCR | HAdV cPCR | GenBank Accession no. (HAdV) | % Nucleotide Identity |
|---|---|---|---|---|---|---|---|
| 1 | CB | 1 | Negative | Negative | Negative | ||
| 2 | MMP | 1 | POSITIVE | POSITIVE | POSITIVE | PP341455 | 99.30 |
| 3 | KSP | 1 | POSITIVE | POSITIVE | POSITIVE | ||
| 4 | MP | 1 | POSITIVE | POSITIVE | POSITIVE | PP341456 | 98.31 |
| 5 | CB | 2 | Negative | Negative | Negative | ||
| 6 | MMP | 2 | Negative | POSITIVE | POSITIVE | PP341457 | 98.92 |
| 7 | KSP | 2 | POSITIVE | POSITIVE | POSITIVE | PP341458 | 98.85 |
| 8 | MP | 2 | Negative | POSITIVE | POSITIVE | ||
| 9 | CB | 3 | POSITIVE | POSITIVE | POSITIVE | PP341459 | 99.01 |
| 10 | MMP | 3 | POSITIVE | POSITIVE | POSITIVE | PP341460 | 98.62 |
| 11 | KSP | 3 | POSITIVE | POSITIVE | POSITIVE | PP341461 | 99.53 |
| 12 | MP | 3 | POSITIVE | POSITIVE | POSITIVE | ||
| 13 | CB | 4 | POSITIVE | POSITIVE | POSITIVE | ||
| 14 | MMP | 4 | POSITIVE | POSITIVE | POSITIVE | PP341462 | 98.48 |
| 15 | KSP | 4 | POSITIVE | POSITIVE | POSITIVE | PP341463 | 99.03 |
| 16 | MP | 4 | Negative | POSITIVE | POSITIVE | ||
| 17 | CB | 5 | POSITIVE | POSITIVE | POSITIVE | ||
| 18 | MMP | 5 | POSITIVE | POSITIVE | POSITIVE | PP341464 | 99.03 |
| 19 | KSP | 5 | Negative | POSITIVE | POSITIVE | PP341465 | 98.68 |
| 20 | MS | 5 | POSITIVE | POSITIVE | POSITIVE |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saasa, N.; M’kandawire, E.; Ndebe, J.; Mwenda, M.; Chimpukutu, F.; Mukubesa, A.N.; Njobvu, F.; Shempela, D.M.; Sikalima, J.; Chiyesu, C.; et al. Detection of Human Adenovirus and Rotavirus in Wastewater in Lusaka, Zambia: Demonstrating the Utility of Environmental Surveillance for the Community. Pathogens 2024, 13, 486. https://doi.org/10.3390/pathogens13060486
Saasa N, M’kandawire E, Ndebe J, Mwenda M, Chimpukutu F, Mukubesa AN, Njobvu F, Shempela DM, Sikalima J, Chiyesu C, et al. Detection of Human Adenovirus and Rotavirus in Wastewater in Lusaka, Zambia: Demonstrating the Utility of Environmental Surveillance for the Community. Pathogens. 2024; 13(6):486. https://doi.org/10.3390/pathogens13060486
Chicago/Turabian StyleSaasa, Ngonda, Ethel M’kandawire, Joseph Ndebe, Mulenga Mwenda, Fred Chimpukutu, Andrew Nalishuwa Mukubesa, Fred Njobvu, Doreen Mainza Shempela, Jay Sikalima, Carol Chiyesu, and et al. 2024. "Detection of Human Adenovirus and Rotavirus in Wastewater in Lusaka, Zambia: Demonstrating the Utility of Environmental Surveillance for the Community" Pathogens 13, no. 6: 486. https://doi.org/10.3390/pathogens13060486











