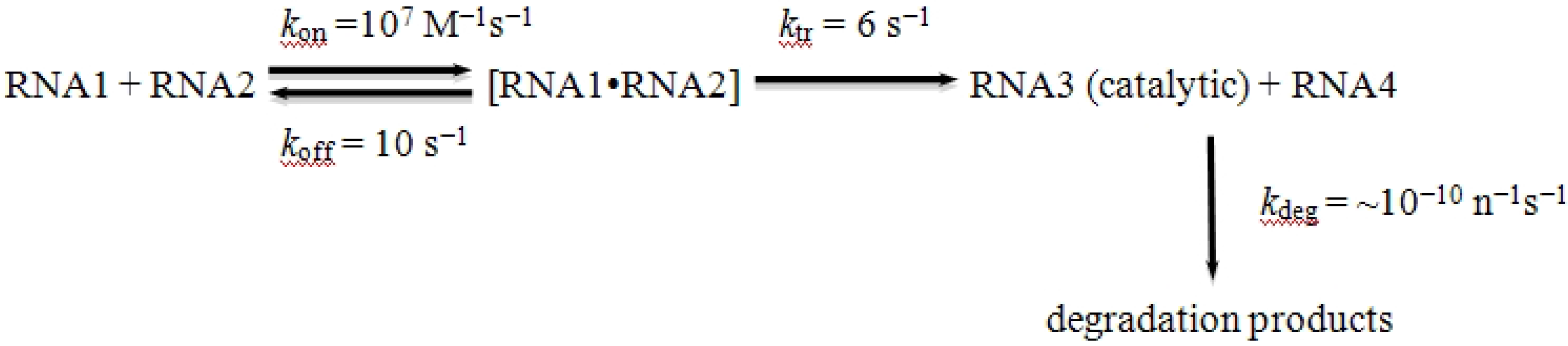1. Introduction
Recombination is a general process in which multi-component entities physically interact and mutually exchange components to create chimeric products, or offspring. While the concept is most often applied to the heredity actions that occur in the living world, parallels exist in chemistry, physics, engineering, and many other scientific disciplines. A simple instance of recombination can be symbolized as:
where
A,
B,
C, and
D represent potentially swappable components, the symbol “•” represents a concrete connection or bond between the components, and
A•B,
etc., represent complete entities with some sort of ascribable value in the system. In the realm of chemical physics for example, one can describe the process of nucleosynthesis as the recombination of existing nuclei to generate new ones. In the realm of biology, recombination of existing genes or genomes can create theretofore novel genotypes. In any event, a characteristic feature of recombination is that relatively large blocks of an underlying material are exchanged among individuals, rather than an alternative process in which new whole entities are made by piecing together minimal subunits, one-at-a-time. If each block contains quantifiable information, then the complexity of the system as a whole can be significantly affected by recombination events.
Clearly life stores and expresses information, and clearly recombination is manifest throughout biology in a variety of forms. Thus its origin and development is of great interest. There have been numerous studies in recombination, approaching such an important phenomenon from different points of view. Nevertheless, recombination is still somewhat of a mystery in terms of its evolutionary origins and maintenance. In this paper we examine the evidence of the early origin of recombination and propose a transition mechanism from an RNA World to cellular life based on recombination processes. We begin by considering contemporary recombination and explore the question of how it might be evolutionarily maintained through a comparison of its advantages and disadvantages on fitness. This section provides the big picture to understand why it is important to understand the reason for the evolutionary origin of recombination, and, whether that reason is still the driving force of the widespread and diverse recombination mechanisms. In the second section we discuss evolutionary pressures that may have played a role in the actual chemical origins of recombination, and which supported possible recombination mechanisms during the RNA World. In the third section we highlight the diversification of the recombination mechanisms with the advent of cellular life. In the last section we discuss the transition of naked genes during the RNA World to genes/genomes during cellular life, in which recombination plays the major role.
1.1. Advantages to Recombination
As early as 1930 Fisher proposed recombination as an advantage for a species because it enables advantageous mutants to become fixed in the population, without having to substitute one from another [
1]. In the absence of recombination two advantageous mutants will only become fixed if they happen in the same lineage, otherwise only one mutant will become fixed while the other will be lost. Later, in 1932 Muller supported this view, and proposed an additional advantage in that recombination also serves as a mechanism to alleviate Muller’s “Ratchet” [
2]. Deleterious mutations occur and accumulate in the population causing a decrease in the mean population fitness. Recombination serves as a mechanism to reconstruct more fit phenotypes from less fit ones in a more prompt manner that back mutations could by themselves, with the addition that more than one solution can be achieved by the shuffling of recombining “good” fragments.
In the 1960s Crow and Kimura [
3] calculated the increase in the rate of substitution of beneficial mutations with and without recombination. They found that for large population sizes of haploid organisms, all beneficial mutants can eventually become fixed by recombination. In the absence of recombination on the other hand, beneficial mutants will only become fixed if they emerge in the same lineage of the most successful mutant; otherwise they will compete and only one will become fixed. In general, the notion is that recombination lowers the “waiting time” for a particular double mutant, but this effect is mainly true in the case of small population sizes [
4,
5]. When large population sizes were considered using deterministic models, recombination was not found to provide a benefit in accelerating evolution [
5,
6,
7]. These results were based on models that did not take into account random drift or linkage disequilibrium. Hill and Robertson [
8] studied the validity of these studies and found that even when there is no initial linkage disequilibrium, nor did the linked genes interact, they still cause interference in one another’s fixation, due to the variance in fitness in the offspring of the mutant compared to the non-mutated form. So each generation the mutants will find a different genetic background or “fitness background”, which will have a differential effect on the frequency that they can reach. Thus, if a mutant lost by random drift reappears by chance, its frequency may not increase or decrease the same as it did before its first removal from the population, due to this differential in genetic background.
Cooper [
9] experimentally demonstrated the advantage of recombination in preventing clonal interference among beneficial mutants, and accelerating evolution. He used populations of
E. coli that lacked the
mutS repair gene, and therefore mutations occur more rapidly. The F plasmid was introduced in the strains to allow for recombination to occur (rec+). Compared to a control rec(–) strain, his experiments show an advantage provided by recombination at high mutation rates, by bringing together beneficial mutants that would otherwise cause mutual interference in each others’ fixation.
Now, if random drift is considered, these models become even more realistic. Random drift generates linkage disequilibrium [
10], which causes interference between linked mutant forms. So, a new mutation that arises physically close to a positively selected mutation will persist for long time, in the absence of recombination, even if the mean fitness is changing from generation to generation. If the new mutation is beneficial as well, their combined effects in increasing mean fitness will be enhanced. In contrast, if the new mutation is slightly deleterious, it will still persist for a long time as a consequence of the linkage, despite the negative effect it has in the mean fitness. In the presence of recombination, the two mutations can be separated, and the slightly deleterious mutant will be more rapidly eliminated by the selective disadvantage of its fitness effect on the mean fitness. This phenomenon will occur more rapidly than it would take for the slightly deleterious mutant to disappear by back mutation. Likewise, the fixation of the advantageous mutant will occur more rapidly if the negative linkage disequilibrium is disrupted.
In summary, recombination seems to provide an advantage to populations (especially small ones) by either reducing competition between advantageous mutants allowing fixation of all or some of them (depending on the effective population size, Ne), or by allowing recovery of the wildtype from deleterious mutants, reducing the effect of Muller’s Ratchet. Beyond these benefits, recombination has other key utilities, which will be discussed later.
1.2. Disadvantages to Recombination
Swapping chunks of genomes is not always beneficial, and can be maladaptive in certain circumstances. In the context of sexual reproduction, the “two-fold cost of sex” is often mentioned as a reason that asexuality persists in nature; parents share at most 50% of identical alleles with their sexual offspring. Another classic case disfavoring genetic swapping is when co-adapted gene complexes are torn apart by recombination. This can be shown mathematically. Kim and Orr found that recombination does not provide an advantage, in a comparison of sexuals and asexuals [
11]. They used a model of DNA sequence evolution in which mutations can be introduced, with differential fitness effects. They specifically compared the probabilities of transition from an advantageous mutant to another, the time lapsed between first and last beneficial mutant substitutions, and the trajectory of the mean fitness values. The three parameters used were found to be similar in both sexual and asexual modes of reproduction, especially when the number of advantageous mutants is not high.
2. Recombination during Pre-Cellular Life: The RNA World
From the above we see that recombination can grant multiple benefits, such as alleviating Muller’s Ratchet, circumventing clonal interference, increasing sequence length, and as a repair mechanism of the genetic information. But while these certainly apply to the maintenance of recombination, it is far less clear which, if any, of these mechanisms serves to explain the value of recombination during initial emergence of life, nor to explain the chemistry of recombination itself. Recombination likely did exist in primeval genomes [
12,
13,
14,
15,
16], and would have been “acquiring” benefits as the genetic machinery became more complex. Thus it is important to consider the advent and early history of strand exchange itself, and consider why it might have been an evolutionary driving force. We begin by focusing on the benefit recombination would have had in building up longer RNA strands from shorter ones during the RNA World, a critical obstacle nascent life would have had to overcome in order to extract complexity out of the informational chaos available from prebiotic chemistry [
17].
2.1. Recombination as a Mechanism to Increase Sequence Length
The RNA World hypothesis, compelling as it is, has some difficulties. First, catalytic RNA (ribozymes) in an RNA World must have been synthesized without templating,
i.e., they had to have originated by a random process. This limits the chemical diversity that could be obtained in the RNA World. The attainable size of a synthetic macromolecule is inherently limited by the repetitive yield or efficiency of the polymerization reaction. The number average size of a polymer population is given by the formula 1/(1–
p), where
p is the probability that any condensing unit has reacted [
18]. This consideration limits chain size considerably, even in the contemporary synthesizer world. If, for example, a non-enzymatic nucleotide (nt) condensation were to have reaction probability of 90–95% (similar to that of montmorillonite-catalyzed condensation [
19]) the product would have an average chain length only of 10–20, which is close to that reported. An alternative condensation method in the water-ice eutectic phase offers the great advantage of requiring lower monomer concentrations, but as of yet has not demonstrated higher reaction probabilities [
20].
The small size limits the chemical repertoire available to a non-templated RNA World. Ribozyme active sites, like those of protein enzymes, generally function as part of a larger structure that holds them in proper alignment. While a Mn
2+-dependent self-cleaving ribozyme has been reported that consists of fewer than 10 nucleotides [
21,
22], other ribozymes are significantly longer. The minimal size of the hammerhead ribozyme is ~20 nt, implying that it would be attainable from an abiotic polymerization, but the polymerase ribozyme studied by Bartel and Unrau [
22,
23,
24,
25,
26] cannot function if it is less than 65–70 nt in length, implying that it would have to appear in a collection of molecules that came out of a synthetic reaction whose efficiency approached 98–99%.
On the other hand, ribozymes cannot be too big. An RNA population of short molecules is necessary to accommodate the maddening tendency of nucleic acids to form stable duplexes. Indeed, contemporary polymerase ribozymes, either natural or selected, show the effects of this “energy trap.” While formation of an individual Watson-Crick base pair results in a relatively low free energy change (ΔG ~ –0.5 kJ mol−1), these small free energies are additive, so that the KD of even a short, 5-bp helix is on the order of 10−6 or more. This leads to the presence of structural traps during RNA refolding that limit the rate at which native, catalytically active structures can be reached.
Any template-dependent polymerization reaction would reach this same kind of trap, and the two strands would not be efficiently separated [
26]. In other words, even small RNAs do not release their partners very efficiently, which makes any template-dependent growth mechanism difficult. Because the rate-limiting step of a ribozyme’s kinetic mechanism, like that of most enzymes, is product release, a long double-stranded product-template structure represents a very serious problem.
For all known replicase ribozymes, the release of product from the double-stranded form is rate limiting and very slow. As a result, the synthetic reaction slows down drastically as the number of phosphodiester bonds formed increases. A primitive RNA ligase or polymerase ribozyme would not be likely to synthesize more than a few bonds in its expected lifetime, because they degrade under physiologically relevant conditions, and any of several phosphodiester bond cleavages in the ribozyme would be fatal to catalytic activity. More complex replicase ribozymes would have been more stable, but their increased length would have made them even less likely to dissociate. Thus, finding solutions to the strand displacement problem is a top priority for those seeking to explore the evolutionary history of replicase ribozymes [
26].
To make matters worse, the formation of double-stranded RNAs makes for a structural—as well as an energetic—trap. RNA scientists are fond of teasing their DNA-focused colleagues about the monotonous nature of the double helix. While this monotony would be a selective advantage when choosing a replicating molecular species, as it allows for mechanistically simple replication, an RNA World that was made up of long RNAs with double-helical structures would not have been complex enough to have active sites. Contemporary RNA polymerizing proteins solve this problem with well-evolved helicase subunits, with special domains that force the two strands apart, or with other activities [
27]. Presumably these activities were not available in the prebiotic RNA world, and so it seems likely that RNA interactions did not involve highly stable helices.
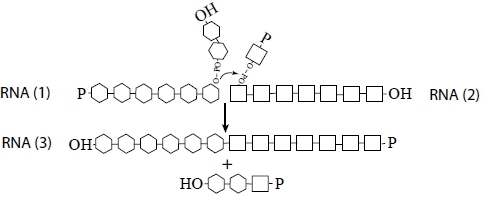
So we are left with a universe of short RNAs, each folded on itself, and all too small to do anything useful. Is there a way out? Can we make an RNA world beyond the oligonucleotide stage? We argue below that recombination, perhaps in conjunction with RNA ligation [
28], supplies a possible answer: a nonreciprocal recombination event between any two RNAs would allow the formation of a species larger than either of the two inputs. The process is be modeled in
Scheme I:
Scheme I.
Kinetics of recombination among RNA polymers.
Scheme I.
Kinetics of recombination among RNA polymers.
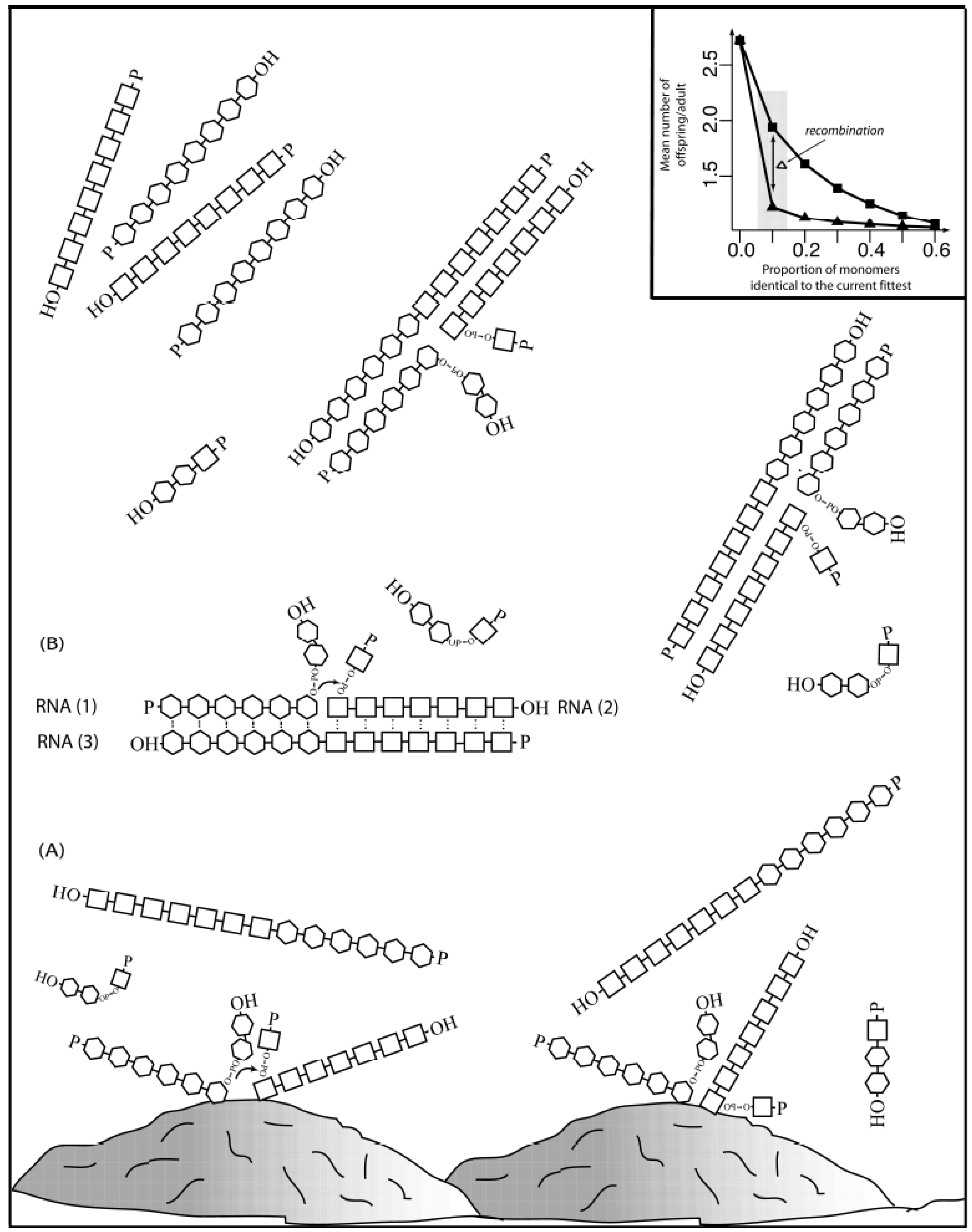
This recombination could be assisted by inorganic surfaces and metal catalysts to counterbalance the intrinsic tendency for RNA oligomers to spontaneously degrade (
Figure 1).
Figure 1 depicts how mineral-aided catalysis (e.g., montmorillonite clay) could have contributed to initiate a recombination event. Random collision of two polymers in the proximity of the clay surface could have facilitated the recombination event (A), giving rise two molecules of different size than the original molecules (
Scheme I). A recombinant molecule, in the presence of a steady state supply of polymers (1) and (2) could have acted as a recombinase (3), in a template-aided (by transient interactions shown as dots) transesterification reaction (shown by an explicit phosphodiester bond with a small arrow from the 3’-hydroxyl to the 5’-α-phosphate). The reaction must happen at a fast enough rate before degradation occurs. The first recombinant molecules in solution could have served in template-driven recombination events (B), increasing the concentration of recombinant molecules in a more exponential way. The inset shows how recombination (bottom line with triangles) could increase diversity to a point in which a population of polymers of 100-monomer units long (for example) could have stayed extant if each parent molecule produced as few as
ca. 1.22 offspring, with up to 10% of the monomers being identical to the most fit. In contrast, the same population in the lack of recombination (upper line with squares) will stay extant only if each parent molecules produce almost twice the amount of offspring,
ca. 1.94.
Figure 1.
Complexity increase through recombination. Inset adapted from [
86] with permission.
Figure 1.
Complexity increase through recombination. Inset adapted from [
86] with permission.
2.3. Constraints on the Generation of an RNA World
The kinetic model for generation of complexity by recombination also constrains the starting conditions for the RNA World. Autocatalysis is an essential feature of living systems, or, presumably, those on the way to life [
34,
35]. If RNA3 in the kinetic mechanism shown in
Scheme I were itself an RNA recombinase, it would appear that it could catalyze its own formation, as long as there was a steady-state concentration of RNAs 1 and 2 (e.g.,
Figure 1). Simulations show a different scenario, however. In simulations, an autocatalytic state is not achieved when the ratio between the initial concentration of the complex between RNA1 and RNA2 and the first-order rate constant for degradation is below ~1 M s
−1 (see
Figure 2 and below). In other words, the finite lifetime of any RNA recombinase means that there is a critical mass of substrate that must be present for it to participate in a self-sustaining autocatalytic process. When the amount of recombining substrate is too low (either through reduced synthesis or enhanced breakdown), autocatalysis can never occur, and the RNA world is doomed before it is born, even if the complex between RNAs 1 and 2 is replenished.
2.4. Biochemistry of RNA Recombination
RNA-directed, templated, recombination of RNA has been observed numerous times, generally involving transesterification caused by ribozymes or protein enzymes (particularly due to strand-switching by polymerases). Non-templated recombination is a different matter: in this case, there must be a transesterification reaction between the two recombining strands.
There are two types of non-replicative
in vitro RNA recombination, as shown by the experiments of Chetverin [
36,
37,
38], with colonies of RNA in grown in agarose media: recombination performed by the RNA itself (spontaneous rearrangements), and recombination dependent on the viral protein enzyme Qβ replicase. Both types of non-replicative RNA recombination occur by transesterification reactions, which differentiate them from template-switching recombination. However, the specific chemical mechanisms of action of the two types of non-replicative recombination vary significantly. The Qβ replicase-dependant RNA recombination requires a “pre-annealing” step. The RNA fragments form first a complex by intermolecular hydrogen bonding; the non-covalent complex facilitate proximity of the recombinants in an attack of the free 3’-OH group of the 5’-fragment to a phosphate group of the sugar phosphate backbone of the 3’-fragment to allow ligation of the recombinant pieces and release of a fragment [
36,
37]. On the other hand, the RNA spontaneous rearrangements occur without participation of the free 3’-hydroxyl group. Presumably it does not require the recombinase enzyme for the reaction. The recombinase could however have a role as a facilitator of the RNA by providing the right environmental conditions for the self-catalyzed splicing reaction [
37].
Chetverin and coworkers’ “molecular colony” or “polony” system allowed for the analysis of rare events that gave rise to replicable RNAs [
36,
37]. Two RNAs, neither of which could be replicated by Qβ RNA polymerase, were allowed to form a complex by formation of a Watson-Crick base-paired region. Recombination was indicated by the formation of an amplifiable RNA, which could be detected by hybridization. Non-templated recombination, as determined by the presence of Qβ-amplifiable RNA species, occurred in solution at a rate of ~10
−9 nt
−1 h
−1. This result should be confirmed when the presence of recombinant RNAs from reaction in solution are assayed by deep sequencing.
The general chemical mechanism of nucleotide polymerization and transesterification involves two Mg
2+ that coordinate to the phosphates and oxygens on either side of the exchanging nucleotides and facilitate the nucleophilic attack of oxygens to form a penta-coordinate intermediate that then resolves to the final product [
38]. If recombination is carried out by transesterification, then it should be Mg
2+-dependent (which it is) and proceed with inversion of configuration, which has not been tested. Chetverin [
39,
40] suggested that a 2’,3’-cyclic phosphodiester would be an essential intermediate of such a process. The mechanism would proceed through formation of the cyclic nucleotide on one strand, followed by attack of the newly released 5’ hydroxyl released on the other strand. This mechanism has some empirical support [
41]; however, the molecular colony [
37] technique should be able to confirm this prediction.
Because RNA itself seems to have a recombinogenic nature and the recombinant product can further replicate, this mechanism seems simplistic enough to argue for its early evolutionary origin, during the RNA World [
12,
15]. RNA can in fact self-recombine in the presence of Mg
2+ in μM concentrations [
41,
42]. Near the origins of life, perhaps the self-recombination process occurred via Mg
2+-catalyzed RNA cleavage with formation of 2’,3’-cyclic phosphates and 5’-hydroxy terminal, which are then cross-ligated. In Chetverin’s experiments, no cryptic ribozyme structures were detected to be involved in the reaction mechanism [
37,
39]. The reaction occurs at a very slow rate: 10
–9 per h per internucleotide bond at 37 °C, with pseudo-first order kinetics. However, even at this rate, some new 10
20 recombinant molecules can be produce in a period on the order of decades, and even if few of those recombinant products get introduced into an exogenous genome, they can produce a significant amount of change in it [
39]. According to the measurements of Lai [
43], the frequency of non-homologous recombination
in vitro is comparable to that of homologous recombination
in vivo, and the difference in their frequencies
in vivo may be due to selection pressures against non-homologous recombination [
36]. More than ten studies between the late 80’s and middle 90’s from different laboratories provide support that RNA recombination occurs
in vivo. Non-homologous, non-replicative RNA recombination has been demonstrated to occur in poliovirus [
44]. Also, the fact that spontaneous RNA rearrangements have been demonstrated to occur at physiological conditions [
37] support this idea.
The implication from all this is that spontaneous RNA rearrangements may have played a crucial role during the early evolution of RNA and DNA molecules [
37,
40]. During the RNA World, the formation of RNA colonies in similar fashion as they do in the agarose media might have been the primitive form of non-membrane aided compartmentalization. Molecules in the colonies may have more resemblance to each other than molecules from other colonies (
i.e., different strains). Non-homologous recombination among them and spontaneous rearrangements of the polymer sequences would have allowed fast growth of colonies and formation of more complex structures. Laminar flow in the aqueous environment (e.g., moisturized clays) could have facilitated migration of RNA molecules that superseded the carrying capacity of the colony to other colonies. Recombination of molecules from different colonies served as a further complexity increasing mechanism, such as sexual reproduction does today.
2.5. The Advent of Polymerization
A string of nucleotides increases by adding one nucleotide after another in a sequence-dependent manner, although once a chain has being made, further elongation can be achieve by either adding one nucleotide at the time or by recombining a string to another. Elongation by adding fragments together can be achieved in a more rapid manner than by adding one nucleotide at the time [
12,
15,
33]. This is because, compared to a phosphoanhydride bond, the α,β-phosphodiester bond is kinetically stable, with a
khydrolysis of approximately 10
–10 per minute, at pH 7 and 37 °C in the common aqueous environments. So, the reaction with a 3’-hydroxyl group needs be catalyzed to have a speed that is far from the hydrolysis rate [
45,
46]. There is no known biological ribozyme that performs this reaction. However,
in vitro evolution has yielded the construction of various RNA enzymes capable of catalyzing the 3’-hydroxyl reaction with a 5’-triphosphate oligonucleotide. The class I ligase catalyzes this reaction with a rate of enhancement of approximately 10
9 compared to the non-catalyzed reaction [
47,
48]. Improvement of this ligase resulted in an enzyme capable of adding up to 20 nucleotides in successive manner to an RNA primer during a 24 h period [
49,
50]. Although this is a great improvement, the catalytic rate is not yet high enough to account for the formation of an RNA molecule of approximately 200 nucleotides. However, because ligation chemistry is the same as that of polymerization chemistry, these ligase ribozymes portend the existence at some point in time of RNA-catalyzed monomer addition, and eventually replicase ribozymes [
51].
Other RNA enzymes evolved
in vitro that are capable of performing this reaction are the hc ligase [
52] which is different in structure and sequence from the class I ligase [
53], and the L1 and R3 ligases, which perform the reaction using an internal template only [
54,
55]. The evolution
in vitro of these various RNA enzymes indicates that perhaps a deeper evolutionary search may uncover a biological RNA enzyme able to perform such an important biological function, related to the origins of life. As shown by
in vitro experimentation, RNA enzymes can perform ligation and polymerization reactions at low speed and with significant lost of fidelity. If fragments were linked together instead of adding mononucleotides one at the time, the energetic investment would have been greatly reduced and the error introduction could have been minimized as well. But at some point the pressure of Darwinian-type evolution for selfishness took over the driving force for replication, and the recombinase mode of replication was superseded by polymerase enzymes [
51].
3. Recombination in Cellular Life
The development of life on the Earth included the establishment of not only polymerization but of compartmentalization into cells. The use of protein enzymes and the entrenchment of the genetic code became prevalent in life. Nevertheless, the utility of recombination persisted, and was presumably co-opted by protein enzymes for a variety of cellular functions. There are some examples of recombination mechanisms that are very minimalistic, which perhaps constitute vestiges of early recombination systems. These include a type of site-specific recombination termed conservative site-specific recombination (CSSR), and the cell-free Qβ RNA recombination system described above. The CSSR is characterized by its simplicity. In this type of recombination, the total number of phosphodiester bonds stays constant through the reaction, and it does not require high-energy cofactors [
56,
57]. This type of recombination is popular in prokaryotes and yeast, but not in higher eukaryotes. In CSSR a four-strand cleavage occurs with the aid of four recombinase monomers that perform the catalytic reaction. Two strands can be cleaved and exchanged before the other two strands are excised and exchanged (λ Int family), or the four strands are cleaved before the exchange events occur (invertase/resolvase family). In both types of recombination, a DNA break is followed by transient covalent bond to the recombinase protein that ensures conservation of energy of the phosphodiester bond [
56]. The fact that there are specific sites of recognition (homology sequences) and breakage for the recombinase, helps the process to occur without synthesis or loss of nucleotides.
The protein-DNA intermediates are covalently bound via conserved residues for each family. For the Int, FLP and Cre proteins, the linkage occurs via 3’-phosphotyrosine. These proteins have three conserved amino acids His, Arg, and Tyr in the conserved carboxy-terminal domain, which is believed to be part of the catalytic center. Modification of these amino acids disrupts recombination without affecting binding of the recombinase to the DNA. These residues are highly conserved in the integrase recombinase families as well. For the invertase/resolvase family, broken DNA attachment to the protein is done via a 5’-phosphoserine linkage. Recombinases within this family have a 13% amino acid identity, especially towards the termini where there are strand exchange determinant elements. The serine residue is highly conserved and its modification causes disruption of the recombination event, without affecting protein-DNA binding.
The minimal recombination machinery can be accompanied by a series of accessory elements that contribute in strand orientation and reaction directionality via DNA-protein interactions. The strand orientation is facilitated by accessory factors that bind only one of the strands to be cleaved. The reaction directionality is rather independent of the equilibrium constant of the un-catalyzed reaction, and the free energy level of the intermediates is similar, facilitating a flow from free substrates to products [
58]. The attB, lox and FRT are minimal recombination sites, formed by the overlap region and the recombinase (Int, Cre and FLP respectively) binding sites. For the λ Int family, the direction of the reaction is determined by two overlapping sets of proteins [
59]. For lox and FRT it is determined by a non-palindromic nature of the overlap region. Additionally, the formation of these high order protein-DNA interactions prevents the formation of unwanted product, optimizing the reaction [
56].
The exact mechanism of the CSSR is not fully clear. The recombinases of the λ Int family do not require divalent metal ion for catalysis, while the invertase/resolvase family seem to require them to some extend to aid in proper protein conformation attainment. The residues involved in the reaction are not the same for both families, but in general Tyr, Arg, His, and Ser seem to play important roles in the catalytic core, although the specific role of each one has not yet been clearly dissected. The recombination reaction consists mainly of a phosphoryl transfer in which the group is linked to a polynucleotide chain of variable length. The residues at the active site not only participate in the reaction
per se, an in-line nucleophilic substitution, but also in ensuring proper alignment of the substrate [
57]. The thermodynamics of the reaction of transferring phosphoryl groups vary largely but they all seem to have a high kinetic barrier [
60]. The rate of enhancement of the catalyzed reaction is of the order of approximately 10
21, which is achieved by an extremely strong binding of the enzyme to the transition state to lower the energy barrier (
Kd ~10
–26 M) [
61]. In terms of energetics, the total amount of energy require for DNA rearrangements is less compared to the amount of energy require for more complex catalytic activities. The recombinase enzymes do not usually turn over; for this reason ATP-dependant chaperones may be required to aid disassembly of the DNA-protein complex (e.g., the
Mu reaction [
62]).
The Qβ RNA recombination system, in contrast to the CSSR, is an RNA viral RNA recombination system that occurs without DNA intermediates. The experimental work with Qβ
in vitro discussed above has demonstrated that recombination is viable in cell-free systems [
36]. An intermolecular RNA recombination was demonstrated first in the 1960s with poliovirus of related strain that exchange genetic markers [
63,
64]. RNA recombination can occur by one of two mechanisms: (1) the splicing-like mechanism, usually among non-homologous recombinants, with its frequency determined by the RNA concentration itself, and being relatively site-unspecific [
36,
65], or (2) the copy-choice mechanism (template jumping), linked to homologous recombination, and with a frequency dependant on viral replicases [
36]. The Qβ replicase recombination system does not seem to exhibit copy choice capabilities [
36], in contrast to retroviral reverse transcriptase enzymes [
66,
67] and to DNA polymerases during PCR procedures [
68,
69]. The template-switch type of recombination requires a dissociation mechanism for the nascent strand that is base-paired to the template. Mechanisms that allow that separation are degradation of the RNA template by RNAse H activity of the reverse transcriptase enzyme, and/or heat-induce melting of DNA duplexes during PCR [
39]. Both of these mechanisms seem to be rather complex to have been available at the origins of life. Thus, perhaps non-homologous recombination is a more antique mechanism from which homologous recombination emerged later on. Two other reasons may be cited in support of this idea: (1) non-homologous recombination is not restricted to a specific site in the RNA fragments to be recombined, and (2) the frequency of the event depends only on the RNA molecules itself.
Lai proposed that these two mechanisms respond to different needs [
43]. The homologous recombination serves as a mechanism to repair replication errors, while the non-homologous recombination provides a quick mechanism to create new genes from its own genome, from other virus genomes, or from a host cell. Non-homologous recombination on the other hand serves primarily as a mechanism to accelerate the evolutionary process. The splicing-type of recombination is perhaps the base mechanism from which the exon-intron gene structure evolved, and even the modern splicing systems [
70,
71]. Perhaps, during the RNA World, self-splicing type of recombination allowed a burst of RNA molecules to be created. Many of the molecules created could have region of sequence homology, as there were not so many choices at the beginning (e.g., small populations of short RNA molecules). Regions of homology could have allowed the emergence of homologous recombination, which served as a repair mechanism and therefore was favored by selection. A mechanism of strand dissociation could have being provided by temperature fluctuation through desiccation cycles in shallow water ponds.
4. The Thread of Recombination throughout the Evolution of Complexity
Life on the Earth began through as-of-yet unknown chemical transitions. At some point, information became stored in linear polymers, and was subject to evolutionary processes that, among other things, led to the manifestation of increasing complexity: order out of chaos [
17]. In this paper we have outlined some of the reasons for why recombination was a critical part of this process. To be succinct, recombination was indispensible because it allowed a rapid means to construct new complex genotypes from old ones,
and because it simultaneously provided a means to preserve the information stored in these new constructs.
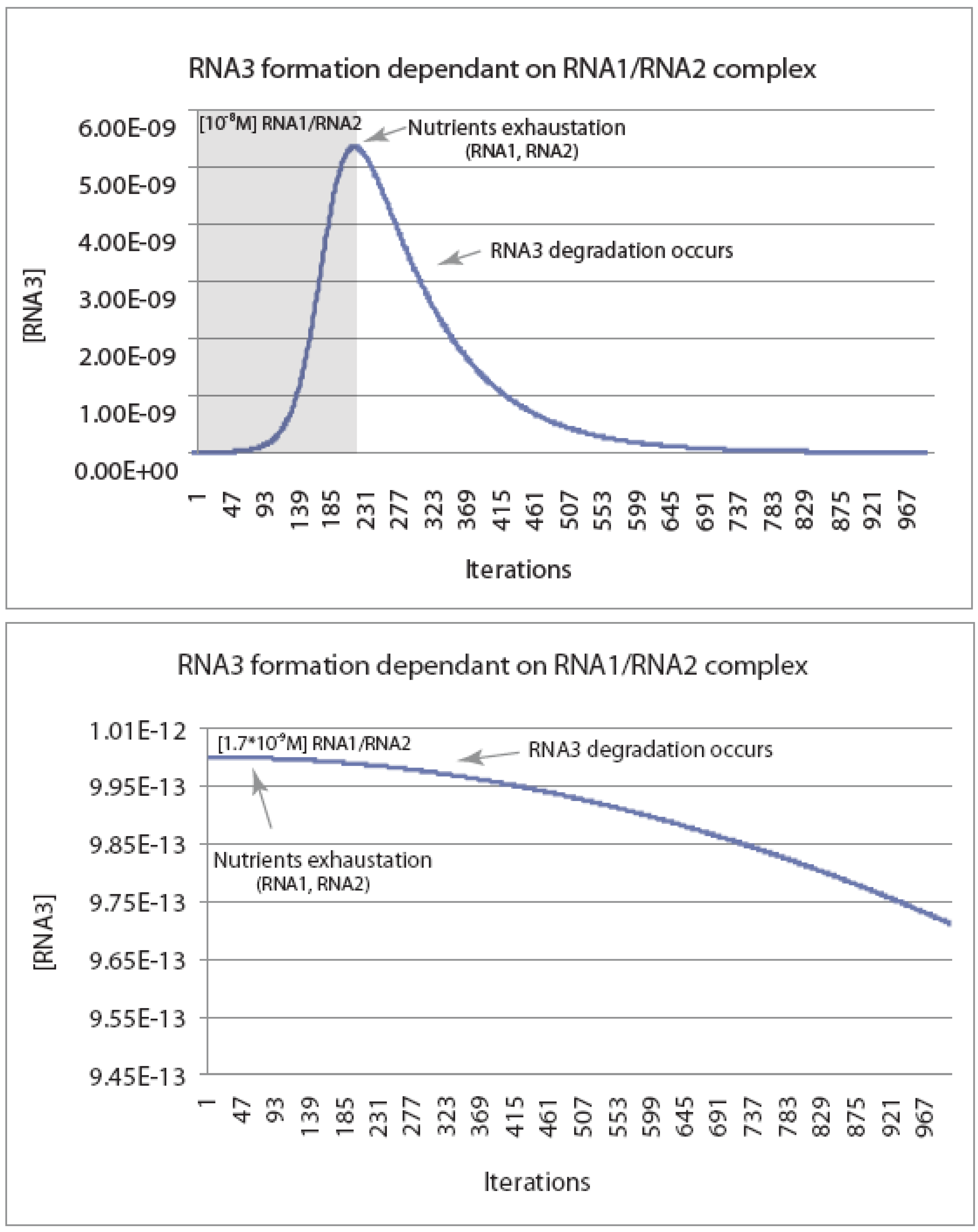
Concerning the former, we have performed a simple simulation iterating RNAs concentrations as a function of time, as expressed in units of 10
5 reaction cycles (
Figure 2). Given estimated RNA breakdown rates, there is a minimal threshold concentration of transiently associated RNAs in the random population at which an autocatalytic cycle could ever occur. To a first approximation, the productive intermediate RNA concentration (RNA1-RNA2) must be greater than about 10
–8 M (or 0.01 μM). This would be a challenge if replication were to start with RNA-catalyzed polymerization, but not necessarily if it were to involve recombination. Primordial RNA-directed RNA polymerization rates would have been no better than the ones discovered to date in the laboratory using quite sophisticated RNA catalysts. For example, the best RNA replicase activity demonstrated to date with a derivative of the class ligase ribozyme requires 5 μM ribozyme and 4 mM of each rNTP to achieve the maximal primer extension seen (20 nt in 24 h) [
24]. This underscores the need for an alternative means of chain lengthening, something that recombination can provide with relative ease [
15,
33].
Figure 2.
Kinetic modeling of the reaction scheme in
Scheme I.
Figure 2.
Kinetic modeling of the reaction scheme in
Scheme I.
Figure 2 shows the concentration of an autocatalytic RNA recombinase (RNA3) as a function of the number of reaction cycles (in units of 10
5 cycles). The model starts with the assumption that RNA3 is a recombinase capable of catalyzing its own formation from RNA1 and RNA2, each of which is present at a frequency of 10
−6 in the random population. Simulations were carried out in Excel using a model where each recombinase could either catalyze the recombination of RNA1 and RNA2 to make more RNA3, or be degraded at one of 10 critical nucleotides per RNA3 species. Simulations were run with the critical parameter being the initial concentration of the intermediate (RNA2-RNA3), which clearly is dependent on the concentration of the random RNA pool. Using the kinetic constants in
Scheme I, the simulations show that the ability to generate autocatalytic expansion of RNA3 is limited by the initial concentrations of RNA1 and RNA2. In the presence of a 10
−8 M [RNA1-RNA2] complex, [RNA3] follows an initial J-shaped growth curve typical of a replicative or autocatalytic process, until “nutrients” RNA1 and RNA2 are exhausted, at which time degradation begins to dominate (top panel). By contrast, when the complex is present at one-sixth this level (1.7 × 10
−9 M), the rate of degradation is large enough so that the recombinase will never increase in concentration (bottom panel). The critical concentration for expansion of the RNA pool is ~10
−8 M, which implies a concentration of the total RNA pool would have to be near 10 mM (recombinase subunits are also assumed to have a frequency of 10
−6 in the random population). Abiotic template-directed replication via a replicase ribozyme may be unlikely to produce initial RNA concentrations this high, suggesting that the RNA population must be surface- or membrane-bound.
Considering the latter, there have been several mechanisms proposed to aid populations of oligomers to keep the genetic information from deterioration during the RNA World:
- (1)
Quasispecies, a model proposed by Eigen [
34], which consisted in a population structure of mutants clouds. The short RNA, or RNA-like, polymers are organized in kinships of different relative frequency fluctuating around a parental “master” sequence of highest frequency. The key feature of this population structure is that selection operates at the quasispecies level and not at the organism level [
72,
73]. This empowers natural selection over random drift and can serve as a route to escape Muller’s Ratchet, as demonstrated experimentally with populations of ligase ribozymes of short length (152 nt) evolved at high mutational rate [
74].
- (2)
Hypercycles (HPC), a model developed by Eigen and Schuster [
75], which consisted in the formation of a cycle of small networks of autocatalytic RNA (or RNA-like) polymers with an altruistic behavior. The product from each autocatalytic network catalyzes the product formation of the adjacent network in the population dynamic. Unfortunately, although the HPC prevent information decay by increasing the rate of product formation, the system itself may be susceptible of being spoiled by “parasitic” mutants—selfish molecules that only care to replicate themselves, disrupting the coupling cycle of “altruistic” networks [
76]. Experimental demonstration of the HPC model on catalytic RNA fragments of short length should be possible and would be of great interest.
- (3)
A model of competitiveness, in which a group of genes could actually act competitively instead of cooperatively, as proposed by the Stochastic Corrector Model (SCM), could have also prevented genetic deterioration. In this model the RNA polymers do not form a coupling cycle of cooperative networks. Instead, the polymers are free to compete and they are replicated by a non-specific replicase [
77,
78]. The SCM and the HPC models could have occurred after an encapsulation process [
76]. If that is the case, genetic deterioration is prevented in a two-way mechanism.
The aforementioned mechanisms could have prevented genetic deterioration during the RNA World, but they all seem too complex to have arisen at the early stage of the RNA World. A more plausible mechanism is that of recombination, because it not only can prevent genetic deterioration (via Muller’s Ratchet), but also it helps to increase sequence length more rapidly. The two benefits can even be contemporaneous, with the additional advantage of the “cheap” energy cost associated with recombination in contrast to polynucleotide synthesis. We have previously proposed that recombination was perhaps the mechanism that helped primeval genomes to increase in sequence length [
12], while others have discussed the limits of recombination in this regard in primitive cellular compartments, but note that recombination can in fact be more ancient that compartmentalization [
14].
Recombination can also serve as a mechanism to prevent genetic deterioration by providing mutational robustness to the population. Szöllösi and Derényi [
79] studied how recombination can generate mutational robustness on networks of genotypes by concentrating them in region of high neutrality of the genotype space. They also evaluated the relationship of the evolution of this robustness with the topology of the network, and considered finite and infinite population models of stem-loop micro RNAs. In their study, the population is considered to be located on the neutral network of the genotypic space. In this network the genotypes have high and similar fitness values. Genotypes located outside the neutral network have lower fitness values and are considered lethal. They showed that although populations of large size tend to be located in region of the neutral network by selection-mutations balance, recombination concentrates the population even further in the neutral network. This effect of recombination, of concentrating the population within the neutral network, was measured by the excess of mutational robustness emerged by the population dynamic (selection-mutation-recombination balance) in comparison with the average neutrality of the network. Thus, similar to its effect on finite populations, recombination provides an increase in mutational robustness given sufficiently polymorphism is already present in the population.
Revisiting Recombination from a Thermodynamic Point of View
Above we pointed out that functional redundancy can be exploited, via recombination, to create functional phenotypes. These phenotypes could be catalytic polymers, in an RNA World, or they could have been novel combinations of gene fragments or even whole genes, in a world of cellular life. A few studies have been done to show the generality of this search process, regardless of the period during the history of life on the Earth in which it took place. Taken together with our arguments, such work secures the thread from life’s origins to its contemporary complexity.
Xia and Levitt [
80] used a 5 × 5 two-dimensional square lattice model of proteins to test for the effects of recombination and mutation in the evolution of protein thermodynamics. They explored all the sequences in the lattice assuming that the protein sequences that fold into the same structure and that differ by a single mutation constituted a neutral network. Mutated sequences survive when they fold into the same structure (global energy minimum) of the prototype sequence (the one with the most robust representation of the optimum structure in the sequence space), if not, they die. The authors found that evolution caused by mutational effects alone is only able to slightly displace the population distribution toward a prototype sequence. This means that many sequences are lost because they would fall in areas of sequence space distant from the prototype sequence. In the case of evolution aided by a combined effect of mutation and recombination, sequences are more likely to fall in sequence space close to the prototype sequence. The ratio of recombination to mutation used was of 1,000:1, enough to bring the population of sequences close to the prototype, despite the large size of the sequence space. The higher the rate of recombination compared to that of mutation, the more compact the sequence space becomes. Thus, recombination induces a mutational robustness because sequences tend to resemble the structure of the prototype sequence, which is the most thermodynamically stable one. In their words, “Recombination acts like a spring that holds sequence population together against the diffusion induced by mutation”. The relationship between increasing the rate of recombination and the consequent observed increase in mutational robustness has also being explained using a two-locus model [
81], and by using gene network models, where negative epistasis was shown to evolve as a byproduct of selection for robustness [
82]. In general, all these studies support the notion that recombination generate genotypes that are within the boundaries of the current mutational cloud and therefore “encourage” the population to stay in the neutral region of the genotypic landscape [
83].
Xia and Levitt went on to propose that when recombination rate is dominant over the (non-zero) mutation rate, the sequences explore more efficiently the vast sequence space and more easily find the prototype sequence in a reasonable amount of evolutionary time, in a flat fitness landscape [
80]. Thus, recombination actually provides some means to solve the paradox that not every genotype or phenotype can be sampled during the history of the universe. Empirically, recombination has been shown to hasten the convergence on a solution to an aptamer selection for binding to a target protein, although given enough time asexual lineages will catch up, as predicted from theory [
84]. Recombination started life, spawned creativity, and then gave way to other mechanisms of replication such as polymerization [
85] but did not disappear from life’s view.