A Novel Multivariate Sample Entropy Algorithm for Modeling Time Series Synchronization
Abstract
:1. Introduction
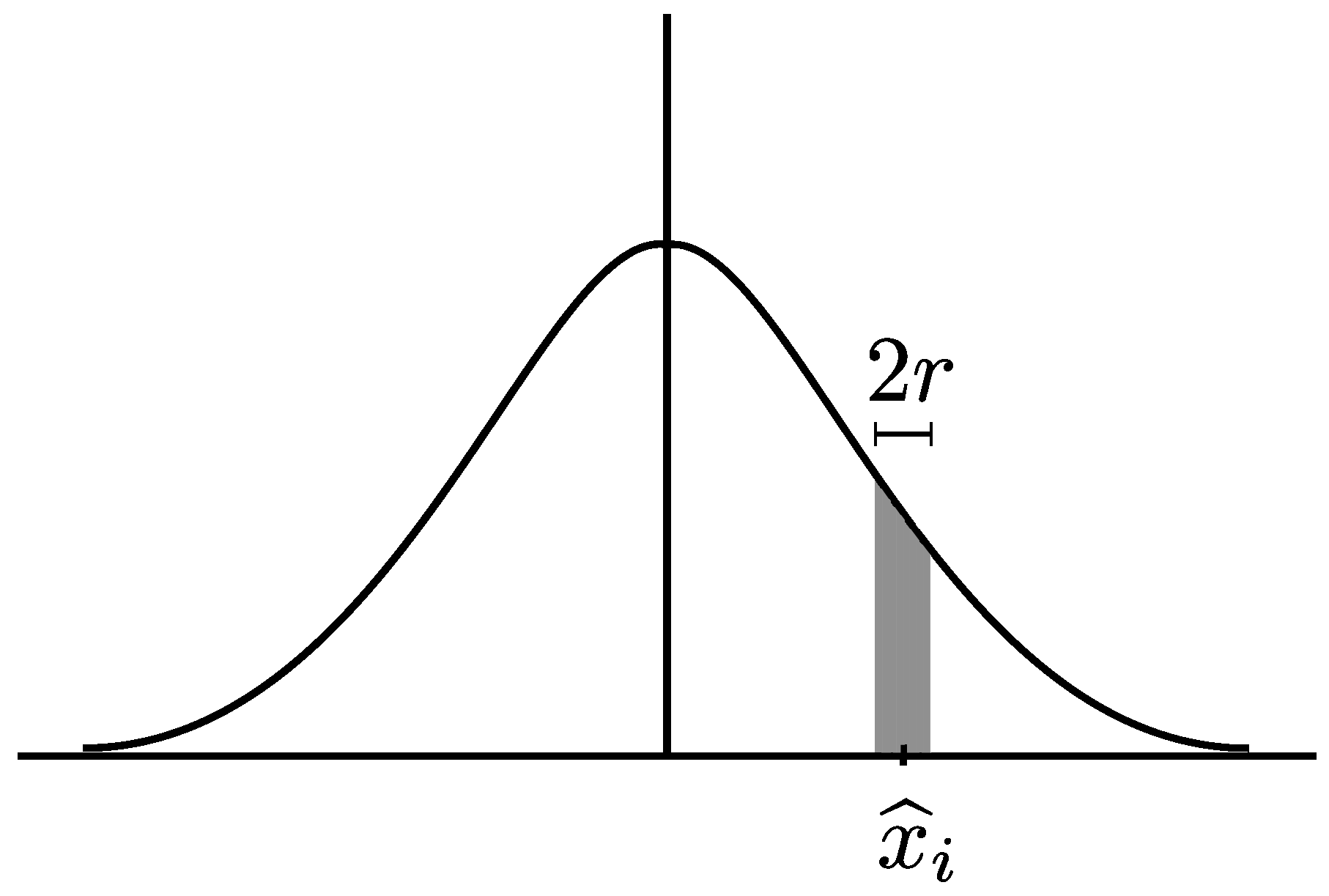
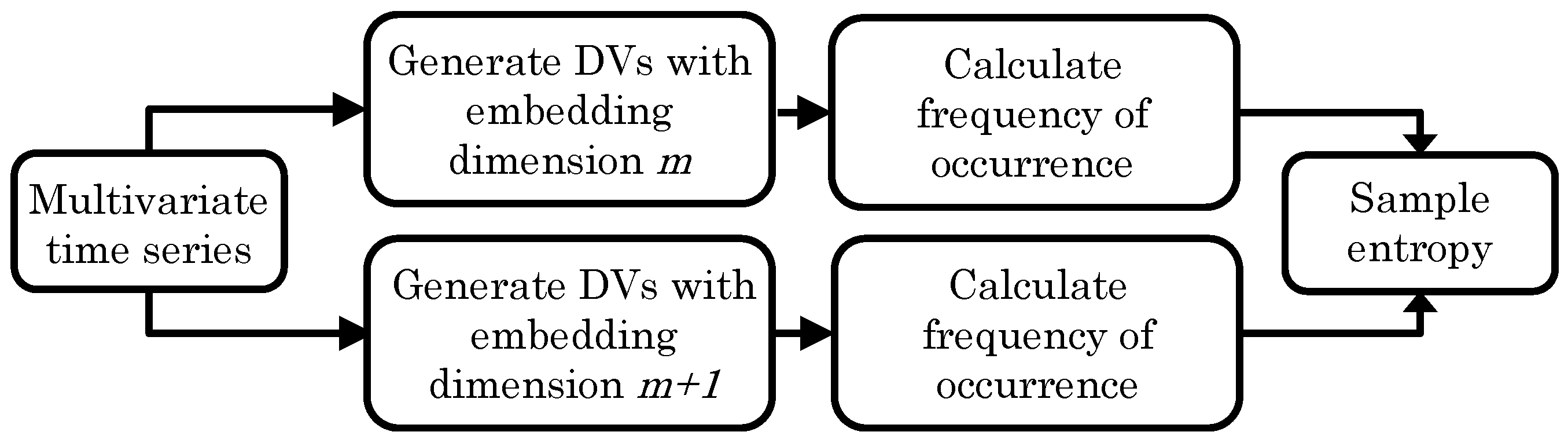
2. Sample Entropy
- For lag and embedding dimension m, generate DVs:where .
- For a given DV, , and a threshold, r, count the number of instances, , for which , , where denotes the maximum norm.
- Define the frequency of occurrence as
- Extend the embedding dimension () of the DVs in step (1), and repeat steps (2) and (3) to obtain .
- The SE is defined as the negative logarithm of the values for different embedding dimensions, that is,
2.1. Multiscale Sample Entropy
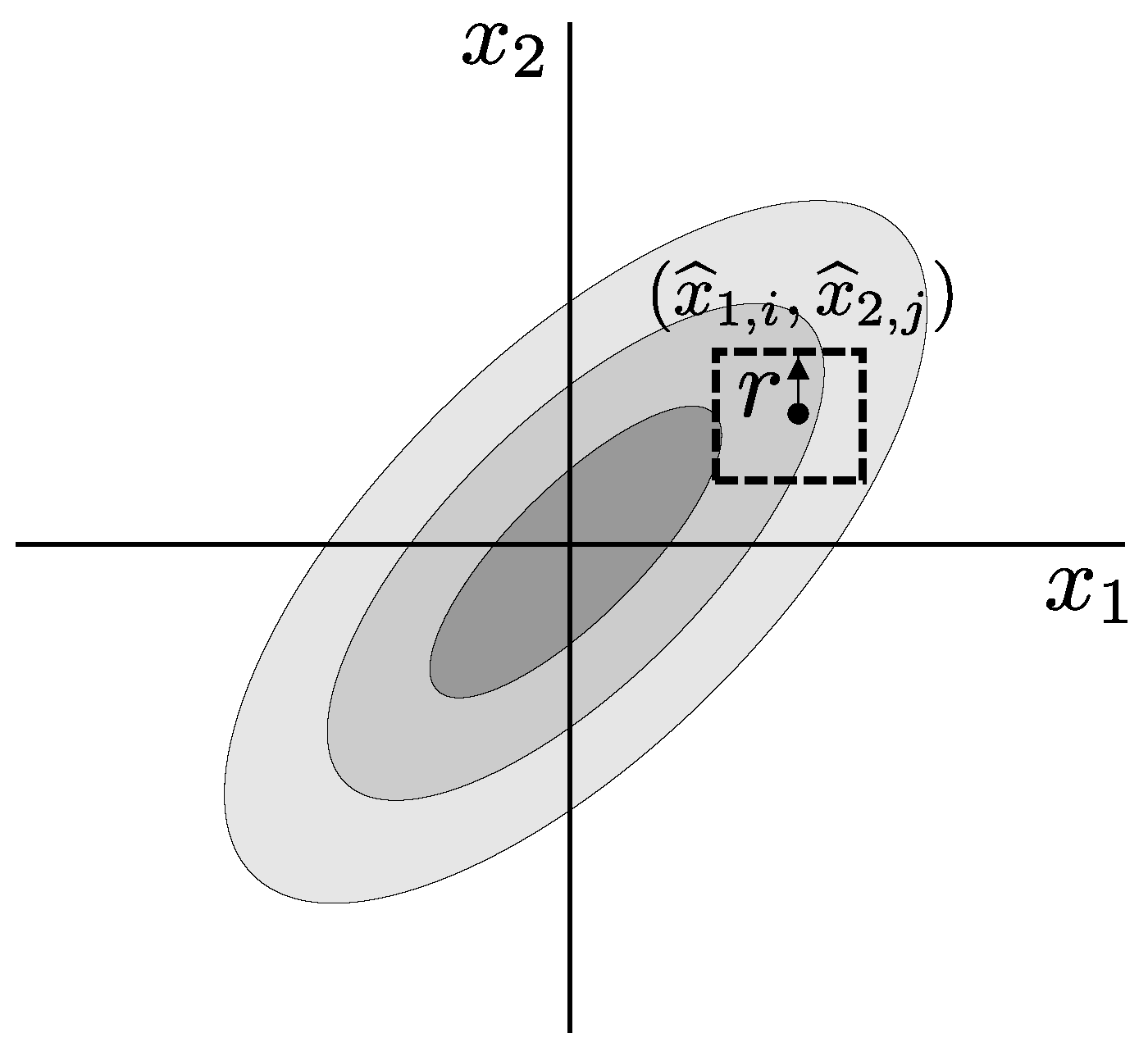
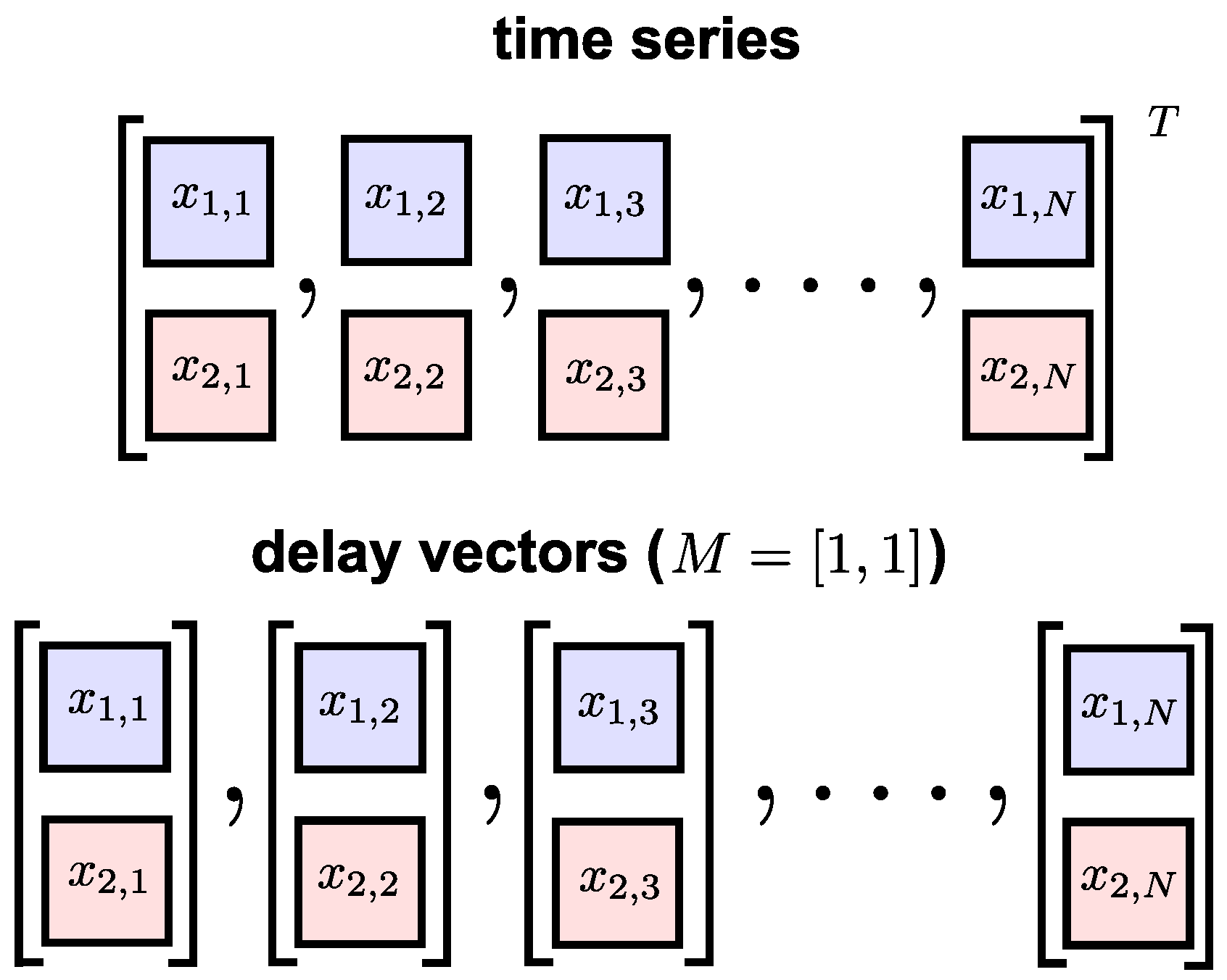
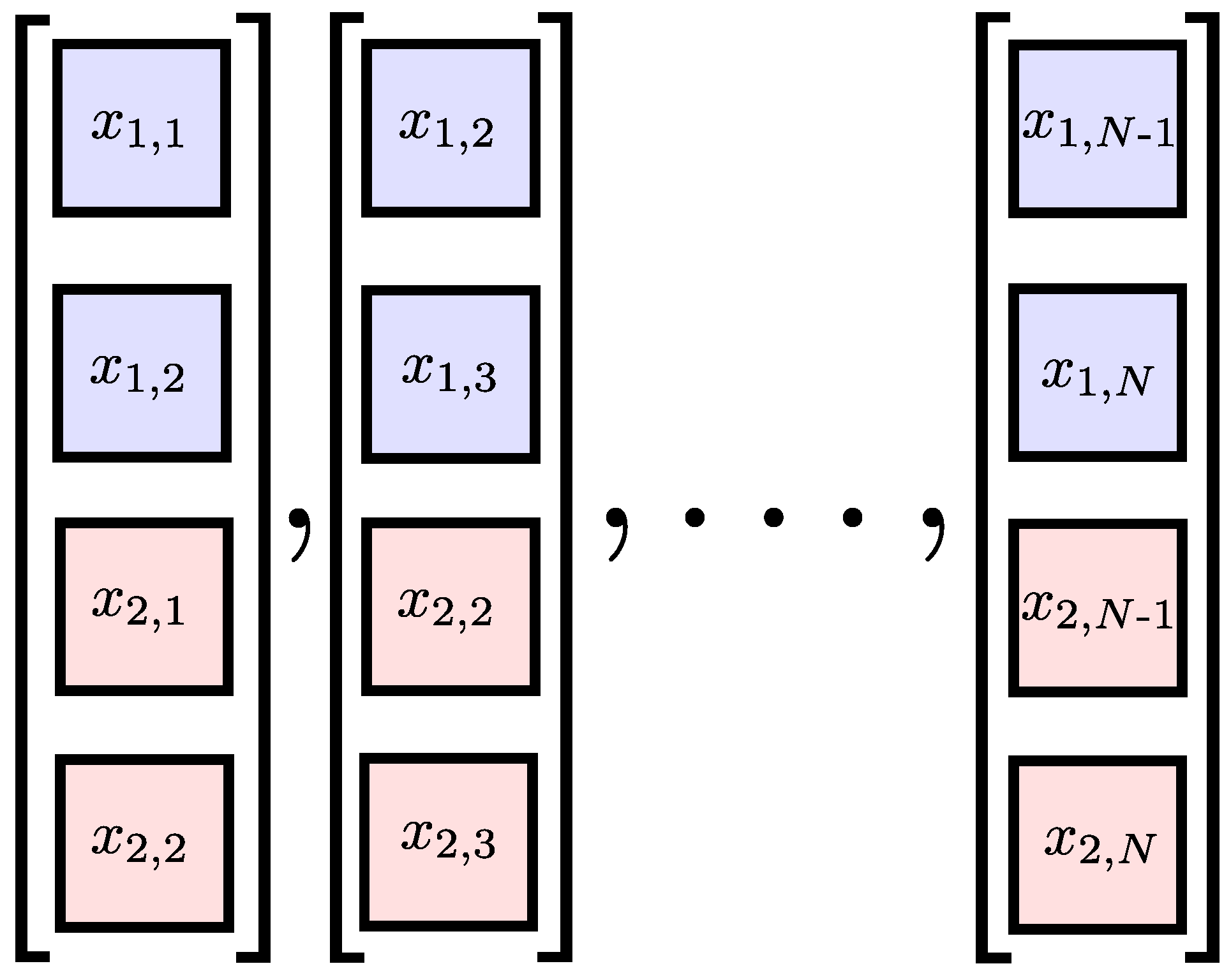
3. Multivariate Sample Entropy
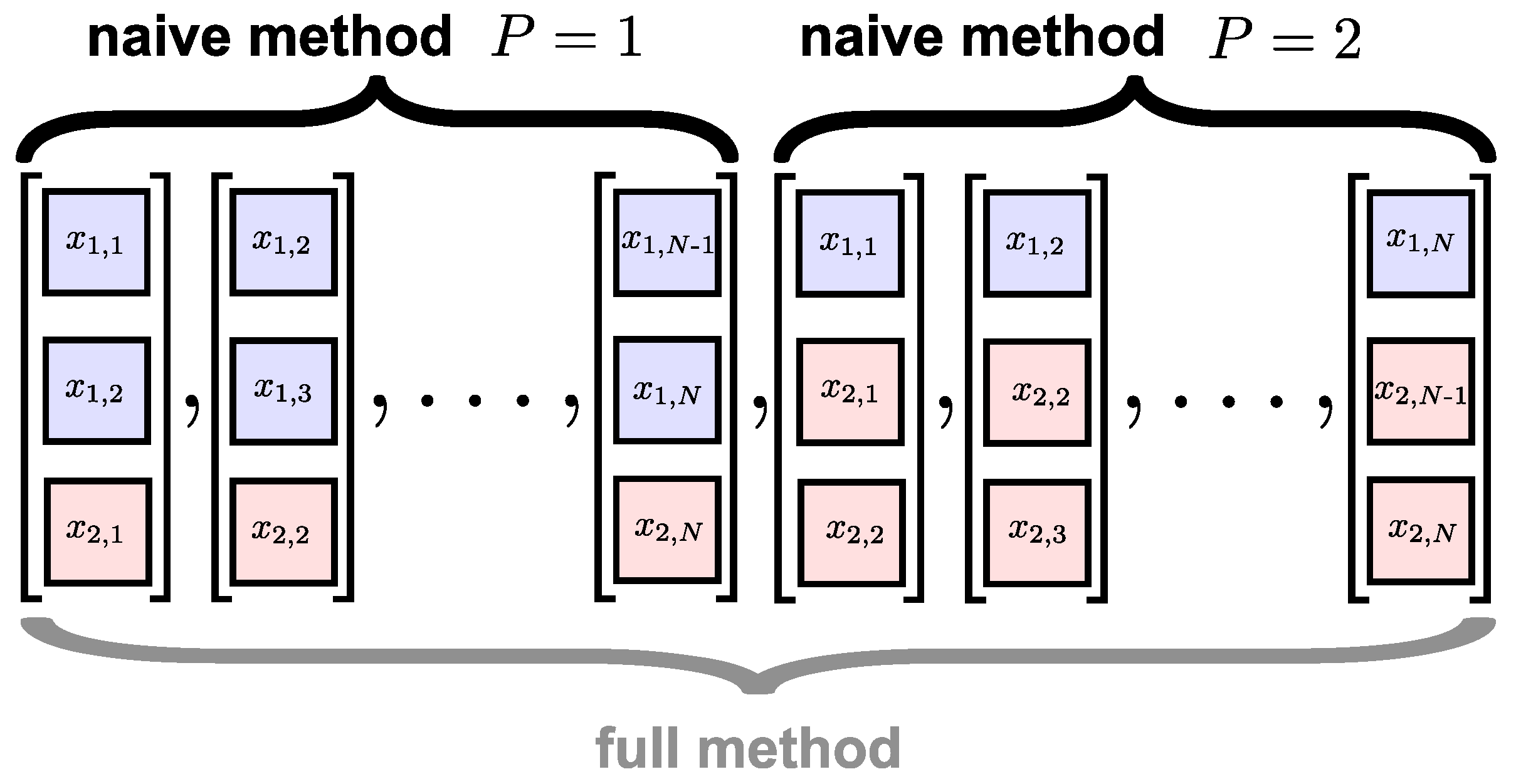
3.1. Existing Algorithms
- Inability to cater adequately for heterogeneous data sources.
- Inconsistencies in the alignment of DVs.
- Counter-intuitive representation of multivariate dynamics.
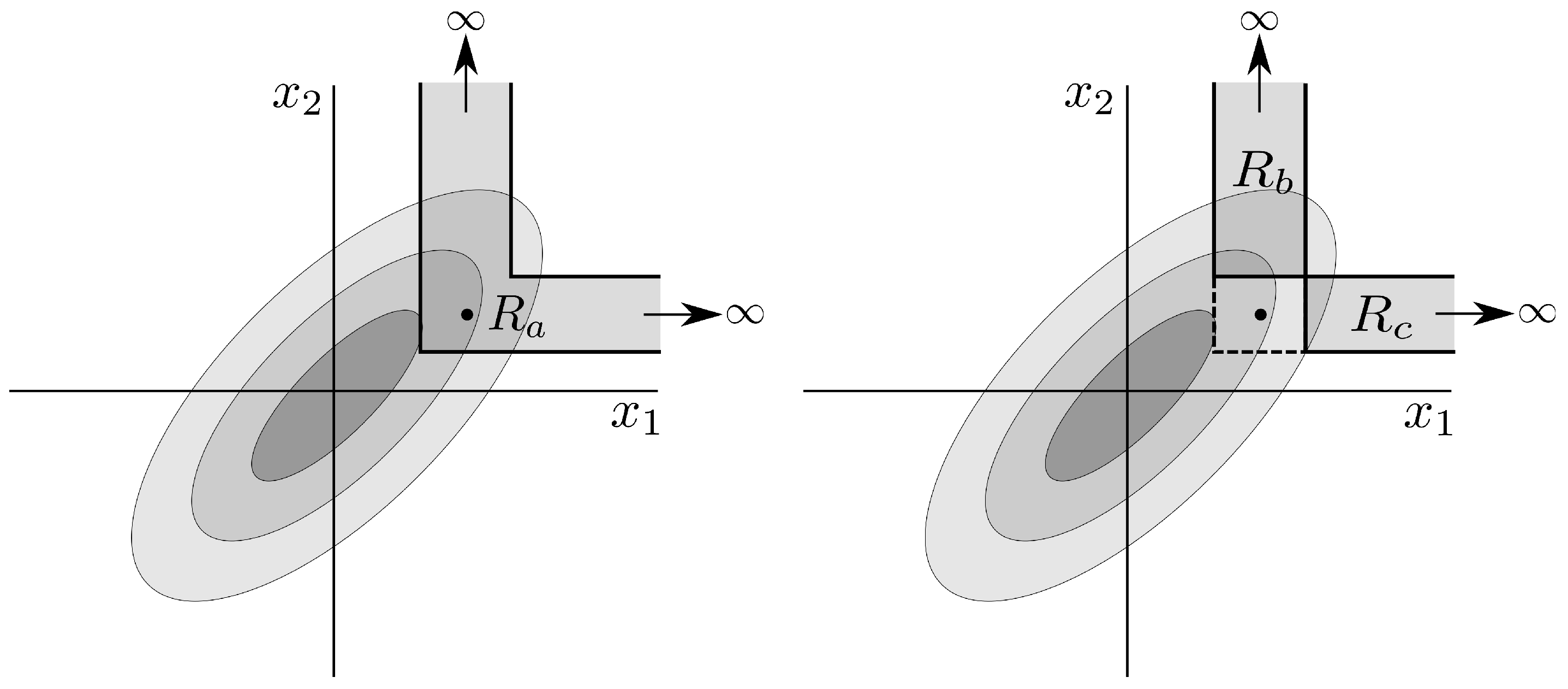
3.2. Synchronized Regularity
3.3. The Proposed Algorithm
3.4. Multivariate Surrogates
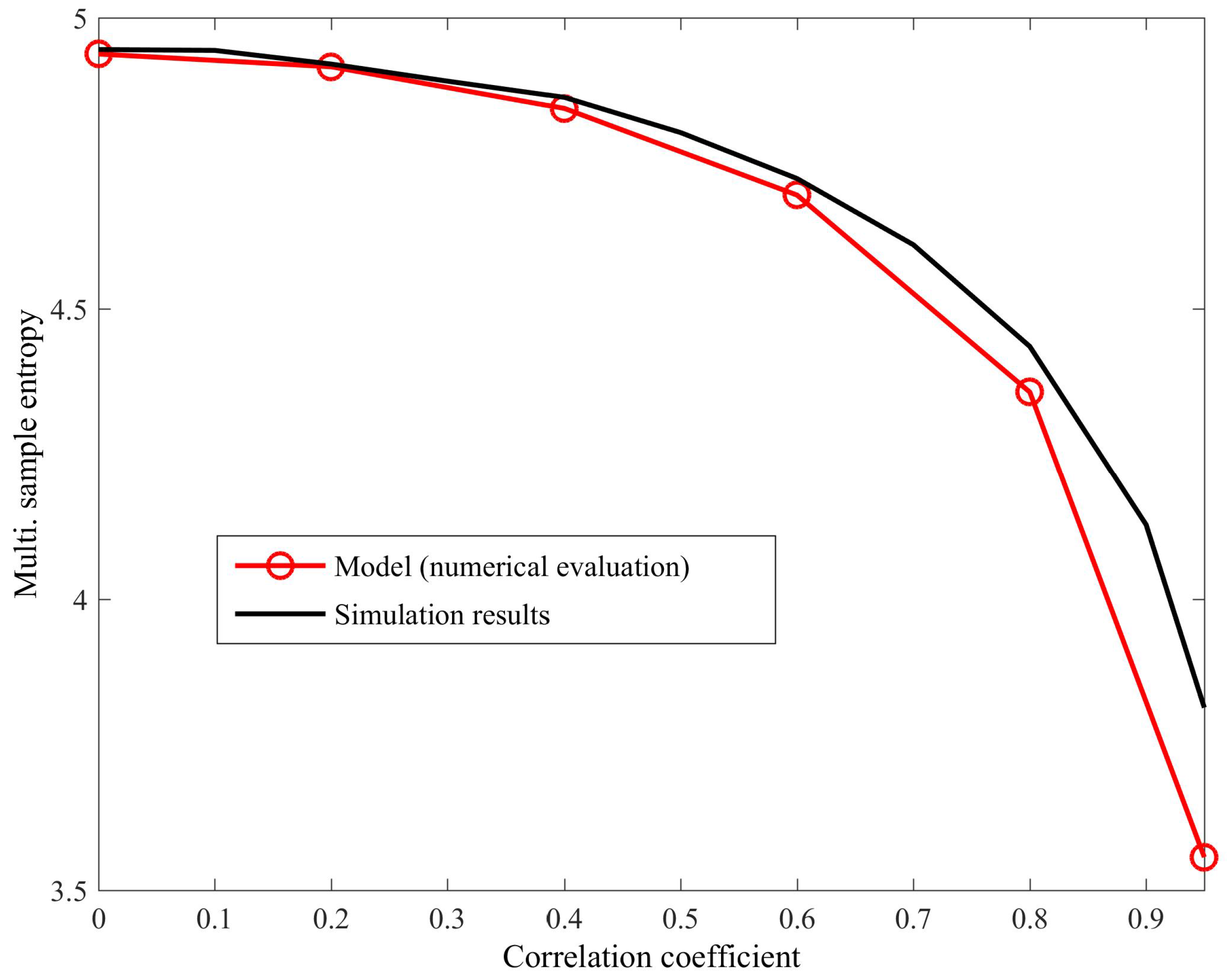
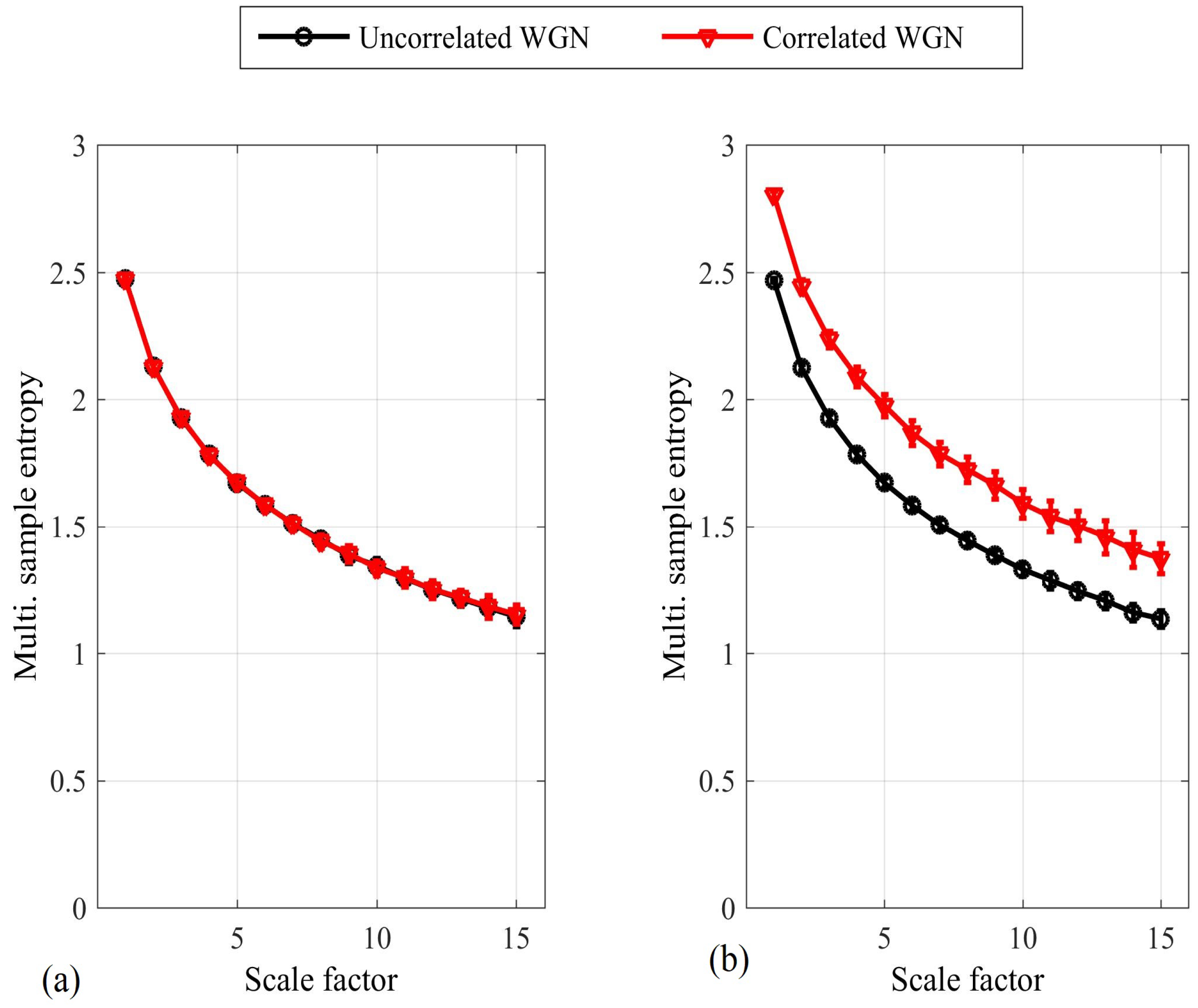
4. Simulations
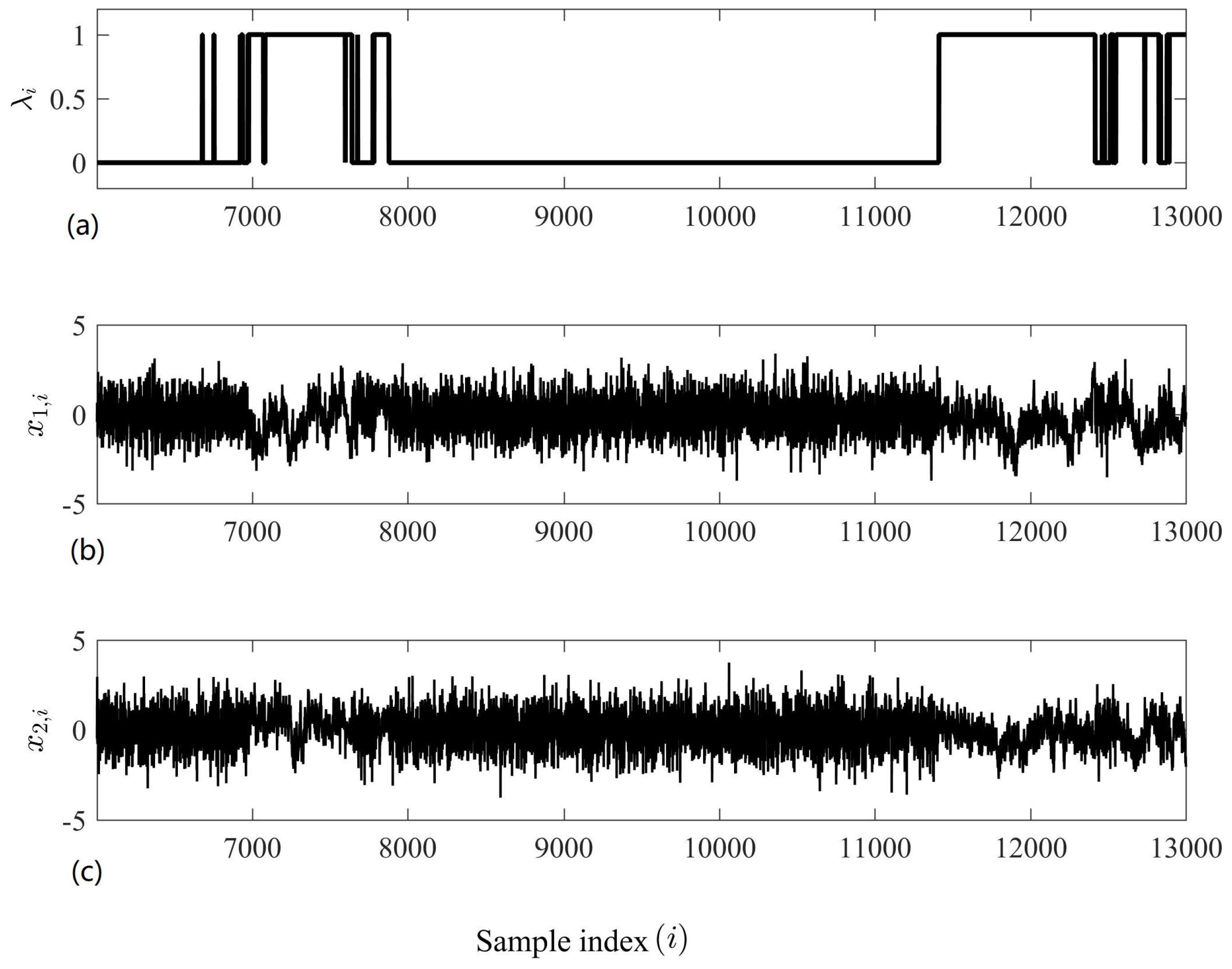
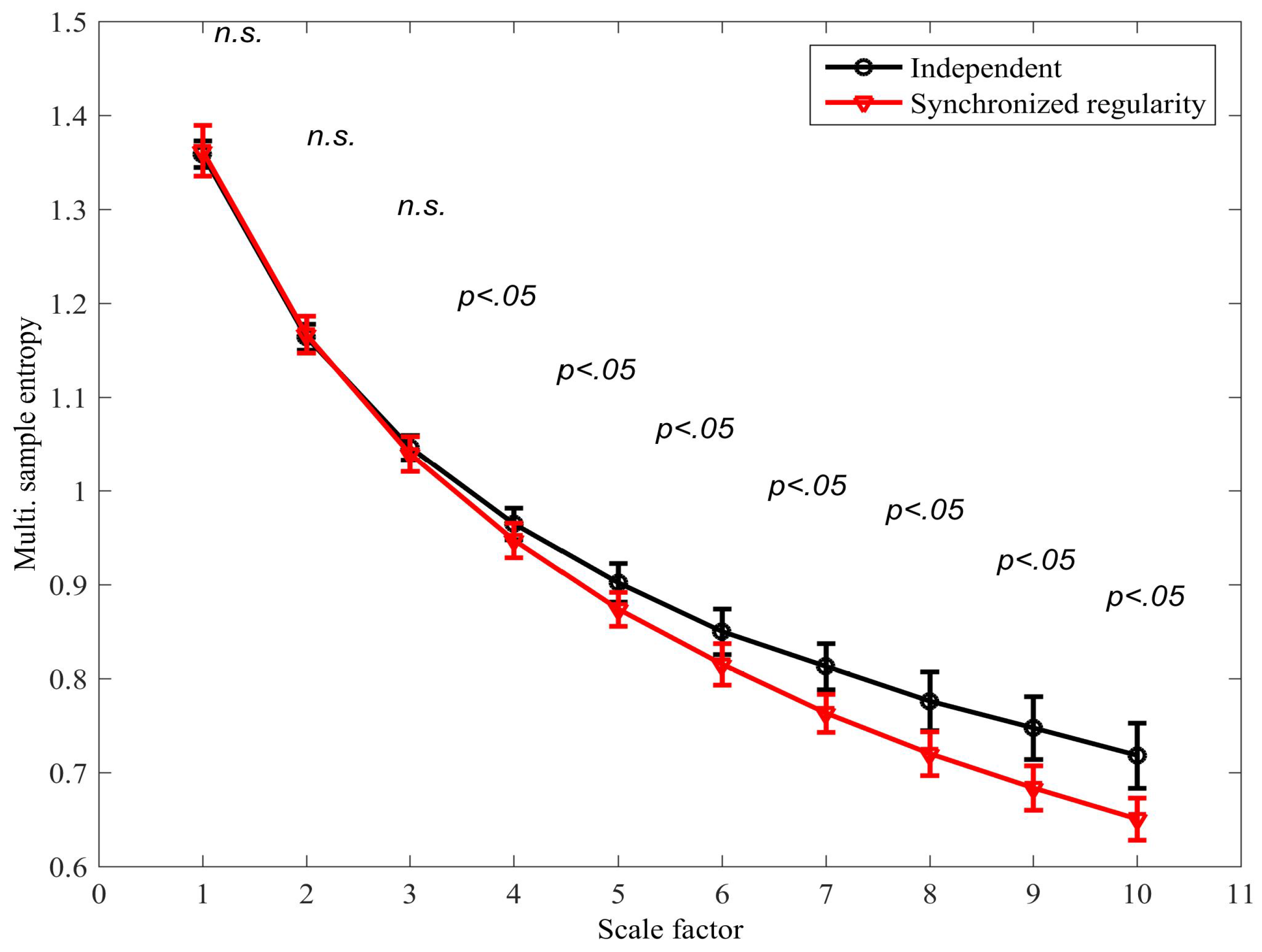
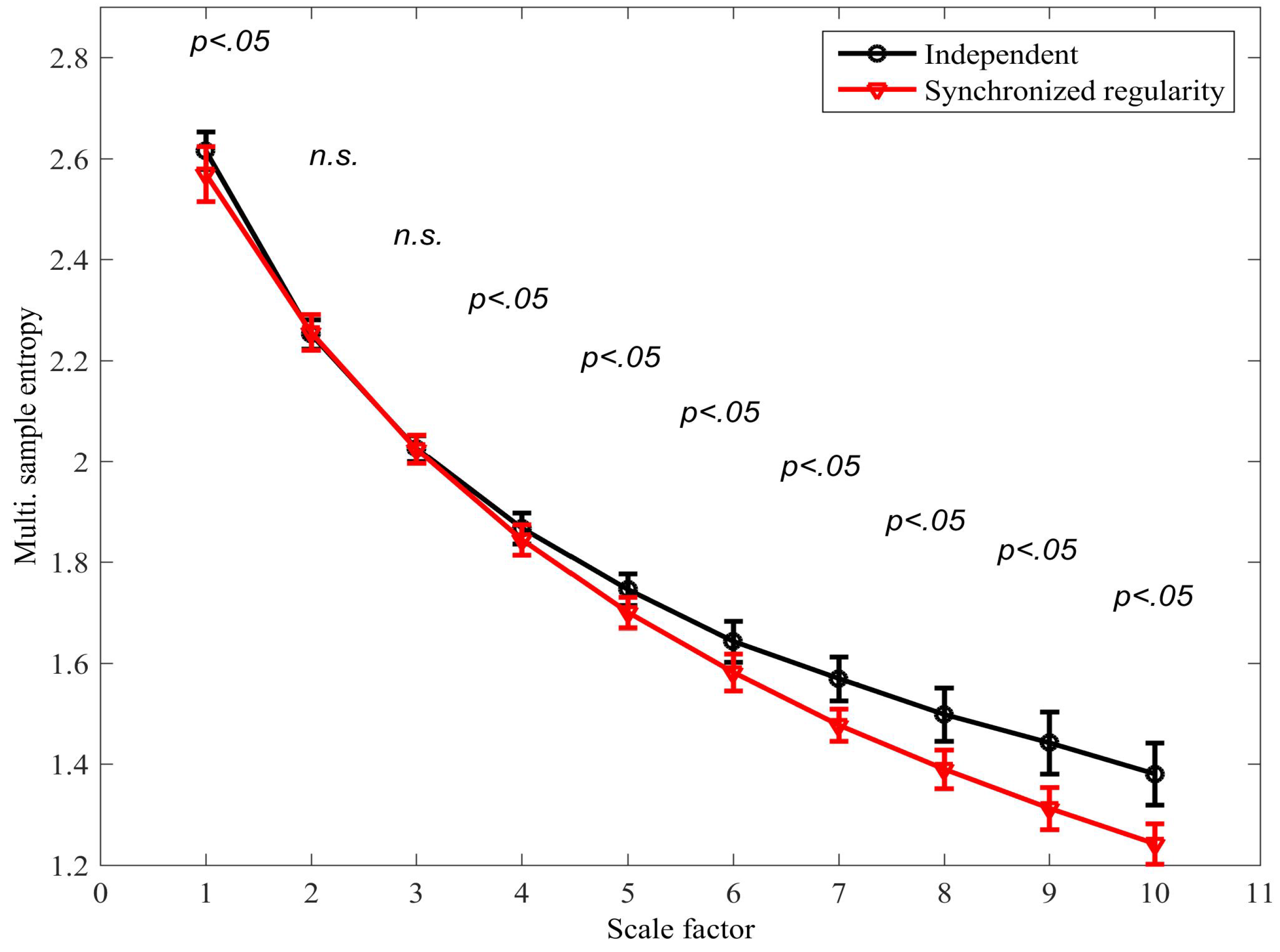
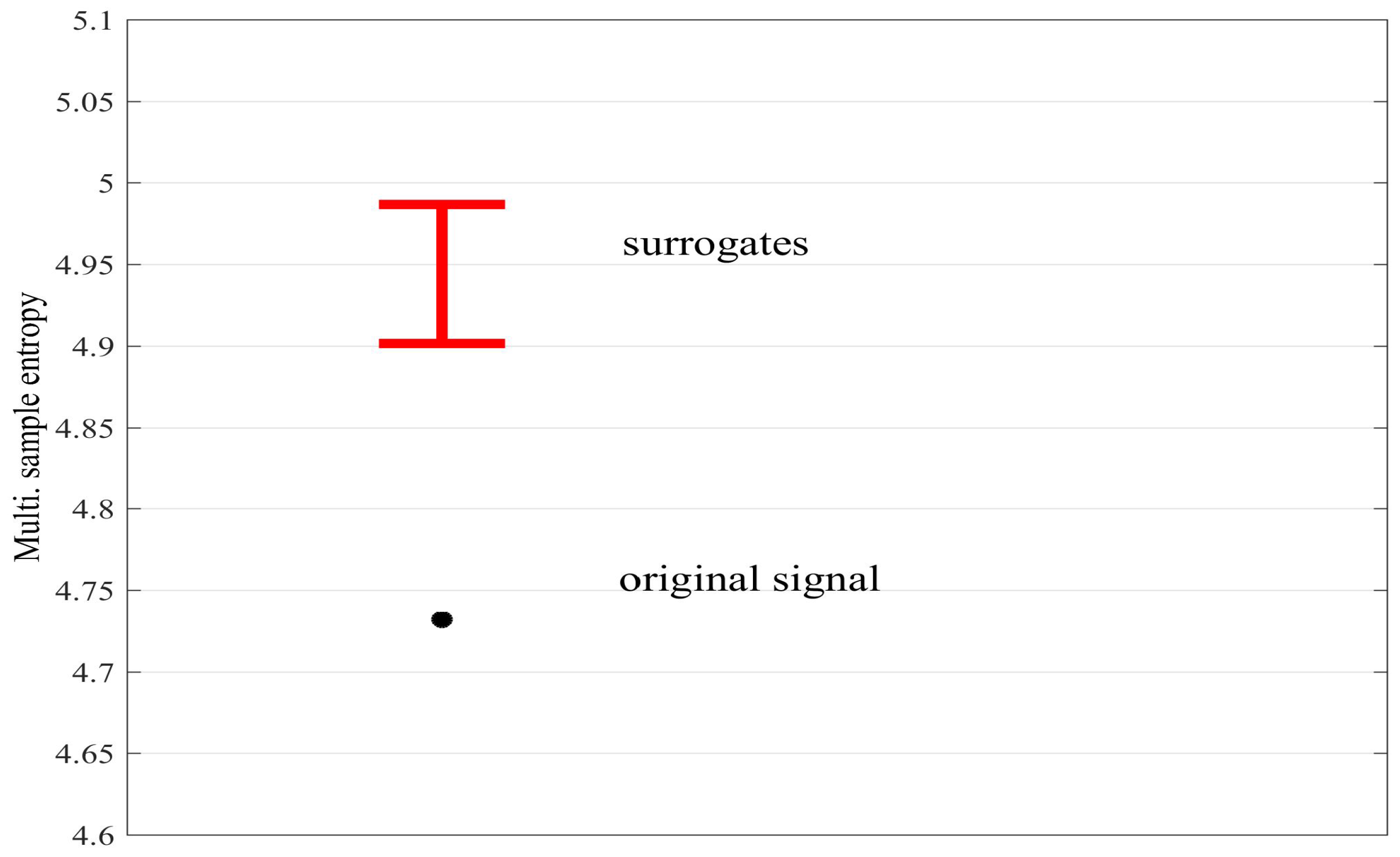
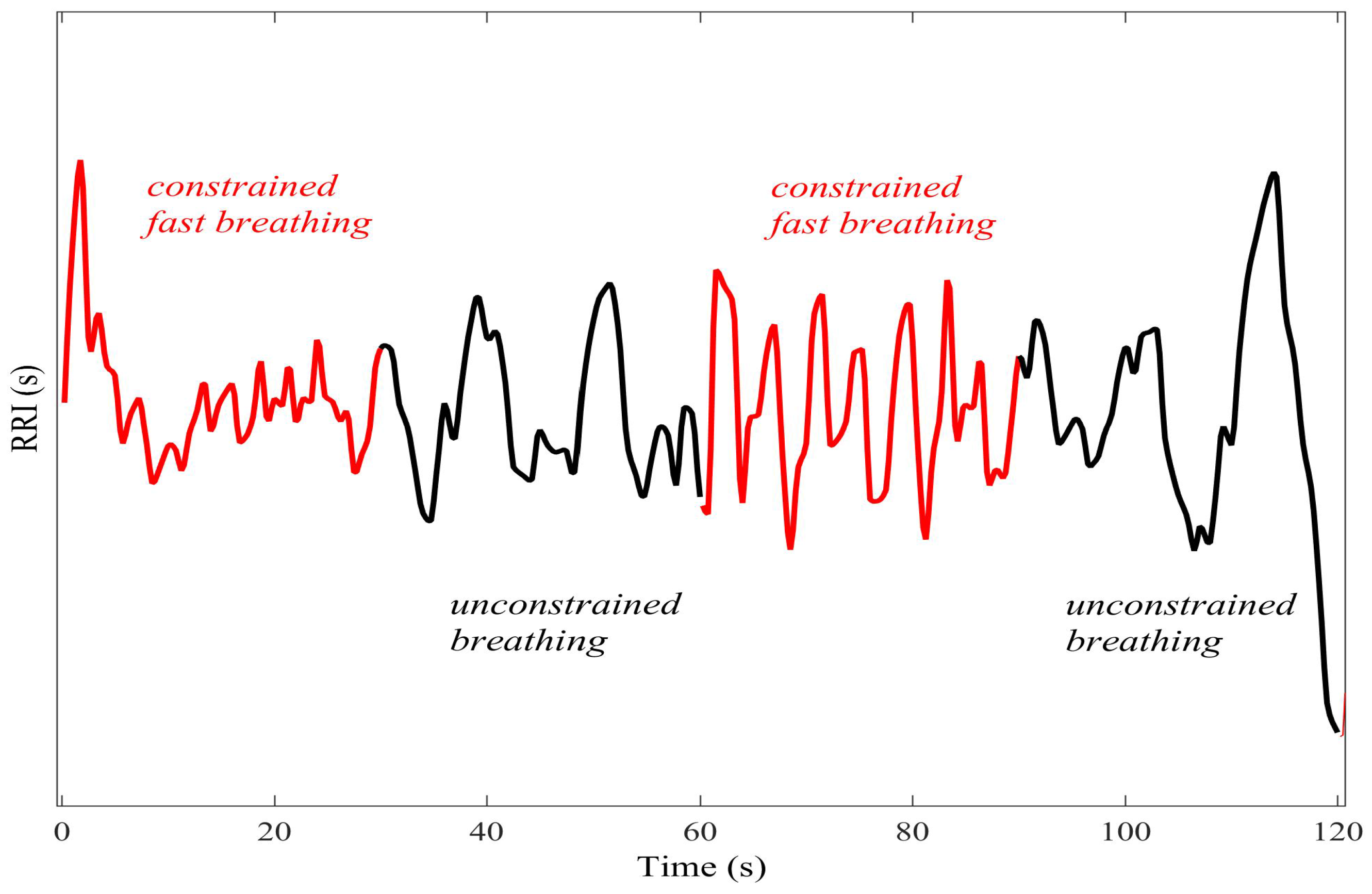
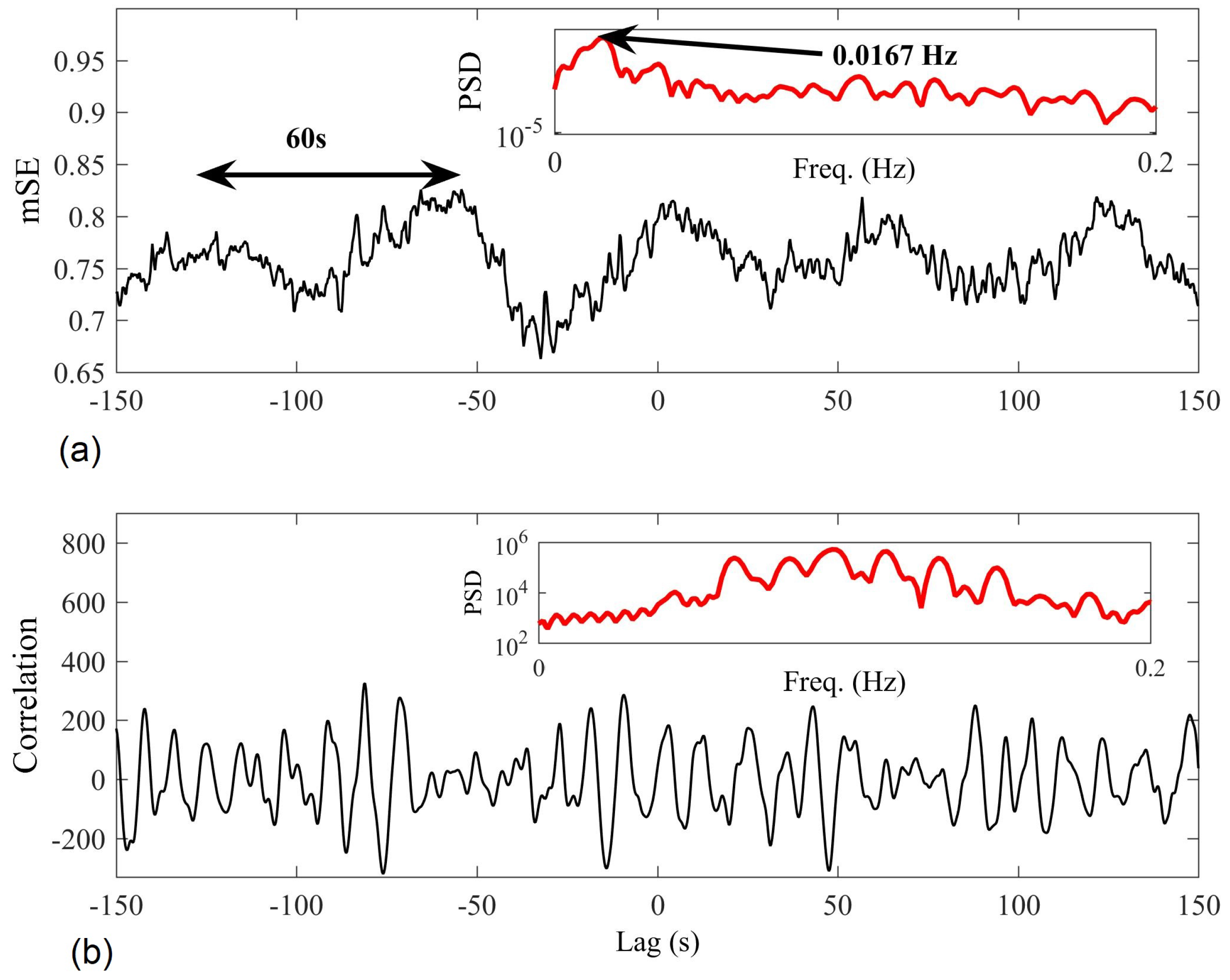
4.1. Detecting Synchronized Cardiac Behaviour
4.2. Classifying Sleep States
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AE | Approximate entropy |
| DV | Delay vector |
| ECG | Electrocardiogram |
| EEG | Electroencephalogram |
| mMSE | Multivariate multiscale sample entropy |
| MSE | Multiscale sample entropy |
| mSE | Multivariate sample entropy |
| RSA | Respiratory sinus arrhythmia |
| SE | Sample entropy |
| WGN | White Gaussian noise |
Appendix A. Multivariate SE for WGN
References
- Pereda, E.; Quiroga, R.Q.; Bhattacharya, J. Nonlinear multivariate analysis of neurophysiological signals. Prog. Neurobiol. 2005, 77, 1–37. [Google Scholar] [CrossRef] [PubMed]
- Burke, D.J.; O’Malley, M.J. A study of principal component analysis applied to spatially distributed wind power. IEEE Trans. Power Syst. 2011, 26, 2084–2092. [Google Scholar] [CrossRef]
- Filis, G.; Degiannakis, S.; Floros, C. Dynamic correlation between stock market and oil prices: The case of oil-importing and oil-exporting countries. Int. Rev. Financ. Anal. 2011, 20, 152–164. [Google Scholar] [CrossRef]
- Granger, C.W.J. Investigating Causal Relations by Econometric Models and Cross-spectral Methods. Econometrica 1969, 37, 424–438. [Google Scholar] [CrossRef]
- Sameshima, K.; Baccalá, L.A. Using partial directed coherence to describe neuronal ensemble interactions. J. Neurosci. Methods 1999, 94, 93–103. [Google Scholar] [CrossRef]
- Schreiber, T. Measuring Information Transfer. Phys. Rev. Lett. 2000, 85, 461–464. [Google Scholar] [CrossRef] [PubMed]
- Pincus, S.M. Approximate entropy as a measure of system complexity. Proc. Natl. Acad. Sci. USA 1991, 88, 2297–2301. [Google Scholar] [CrossRef] [PubMed]
- Richman, J.S.; Moorman, J.R. Physiological time-series analysis using approximate entropy and sample entropy. Am. J. Physiol. Heart Circ. Physiol. 2000, 278, H2039–H2049. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.; Goldberger, A.L.; Peng, C.K. Multiscale entropy analysis of complex physiologic time series. Phys. Rev. Lett. 2002, 89, 068102. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.; Goldberger, A.L.; Peng, C.K. Multiscale entropy analysis of biological signals. Phys. Rev. E 2005, 71, 021906. [Google Scholar] [CrossRef] [PubMed]
- Bruce, E.N.; Bruce, M.C.; Vennelaganti, S. Sample entropy tracks changes in electroencephalogram power spectrum with sleep state and aging. J. Clin. Neurophysiol. 2009, 26, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.; Ghiran, I.; Peng, C.K.; Nicholson-Weller, A.; Goldberger, A.L. Complex dynamics of human red blood cell flickering: Alterations with in vivo aging. Phys. Rev. E 2008, 78, 020901. [Google Scholar] [CrossRef] [PubMed]
- Hornero, R.; Abásolo, D.; Escudero, J.; Gómez, C. Nonlinear analysis of electroencephalogram and magnetoencephalogram recordings in patients with Alzheimer’s disease. Philos. Trans. R. Soc. Lond. A Math. Phys. Eng. Sci. 2009, 367, 317–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, M.U.; Li, L.; Cao, J.; Mandic, D.P. Multivariate multiscale entropy for brain consciousness analysis. In Proceedings of the IEEE Engineering in Medicine and Biology Society (EMBC), Boston, MA, USA, 30 August–3 September 2011; pp. 810–813. [Google Scholar]
- Ahmed, M.U.; Mandic, D.P. Multivariate multiscale entropy: A tool for complexity analysis of multichannel data. Phys. Rev. E 2011, 84, 061918. [Google Scholar] [CrossRef] [PubMed]
- Kaffashi, F.; Foglyano, R.; Wilson, C.G.; Loparo, K.A. The effect of time delay on Approximate & Sample Entropy calculations. Phys. D Nonlinear Phenom. 2008, 237, 3069–3074. [Google Scholar]
- Rosenblum, M.G.; Pikovsky, A.S.; Kurths, J. Phase Synchronization of Chaotic Oscillators. Phys. Rev. Lett. 1996, 76, 1804–1807. [Google Scholar] [CrossRef] [PubMed]
- Rosenblum, M.G.; Pikovsky, A.S. Detecting direction of coupling in interacting oscillators. Phys. Rev. E 2001, 64, 045202. [Google Scholar] [CrossRef] [PubMed]
- Rulkov, N.F.; Sushchik, M.M.; Tsimring, L.S.; Abarbanel, H.D.I. Generalized synchronization of chaos in directionally coupled chaotic systems. Phys. Rev. E 1995, 51, 980–994. [Google Scholar] [CrossRef]
- Cao, L. Dynamics from multivariate time series. Phys. D Nonlinear Phenom. 1998, 121, 75–88. [Google Scholar] [CrossRef]
- Bhattacharya, J.; Pereda, E.; Petsche, H. Effective detection of coupling in short and noisy bivariate data. IEEE Trans. Syst. Man Cyberne. Part B Cybern. 2003, 33, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Netoff, T.I.; Schiff, S.J. Decreased neuronal synchronization during experimental seizures. J. Neurosci. 2002, 22, 7297–7307. [Google Scholar] [PubMed]
- Hayano, J.; Yasuma, F.; Okada, A.; Akiyoshi, M.; Mukai, S.; Fujinami, T. Respiratory Sinus Arrhythmia. Circulation 1996, 94, 842–847. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.R.; Wermuth, N. A simple approximation for bivariate and trivariate normal integrals. Int. Stat. Rev. 1991, 59, 262–269. [Google Scholar] [CrossRef]

| Existing | Proposed | % W | % SWS | |
|---|---|---|---|---|
| Excerpt 1 | 14.5 | 19.9 | ||
| Excerpt 4 | 27.1 | 6.5 | ||
| Excerpt 5 | 10.8 | 28.6 | ||
| Excerpt 6 | 3.3 | 32.3 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Looney, D.; Adjei, T.; Mandic, D.P. A Novel Multivariate Sample Entropy Algorithm for Modeling Time Series Synchronization. Entropy 2018, 20, 82. https://doi.org/10.3390/e20020082
Looney D, Adjei T, Mandic DP. A Novel Multivariate Sample Entropy Algorithm for Modeling Time Series Synchronization. Entropy. 2018; 20(2):82. https://doi.org/10.3390/e20020082
Chicago/Turabian StyleLooney, David, Tricia Adjei, and Danilo P. Mandic. 2018. "A Novel Multivariate Sample Entropy Algorithm for Modeling Time Series Synchronization" Entropy 20, no. 2: 82. https://doi.org/10.3390/e20020082





