A fundamental aspect of modern science is that information is physical [
1]. Information must have an instantiation, and may informally be described as patterns shared between structures. A key, for example, carries information about the lock that it fits, whereas this text is information because it corresponds to presumable patterns in the brains of the author and the reader. More specifically, information is defined as the correlated entropy between two ensembles [
2], and the ambiguous concept of entropy [
3] represents a fundamental link between thermodynamics and information theory [
4].
This link between matter and information is most evidently manifested by the molecular constitution of the genetic code, and represents the core of modern life science. Genetic information, in the form of nucleic acids, is propagated by a thermodynamic mechanism that may be described as template replication of aperiodic polymers [
5,
6], and this surprisingly simple concept combines the replication and variability that underlies Darwinian evolution [
7,
8,
9]. In principle therefore, the origin of life may be explained by the spontaneous rise of template-replicating polymers [
10]. But despite significant advances in the field of autocatalytic and self-replicating chemistry [
11,
12,
13,
14,
15,
16], self-organization of template-replicating polymers from individual monomer, independent of complex catalysts, has not been demonstrated. This inability to physically demonstrate spontaneous rise of genetic information represents a definitive missing link between thermodynamics, information theory, and life science [
17], and has been embraced as a key argument by those who oppose a scientific explanation for life [
18].
In order to meet this challenge we set out to design a system of simple monomers that self-organize into template-replicating polymers, and started by theoretically analyzing the minimal requirements for a system comprising such dynamics. Based on Watson and Crick’s well-defined model of the DNA molecule [
5,
6] we deduced that the concept could be expressed in terms of monomeric objects interacting by two interrelated binding mechanisms responding to environmental fluctuations in energy (
Fig. 1):
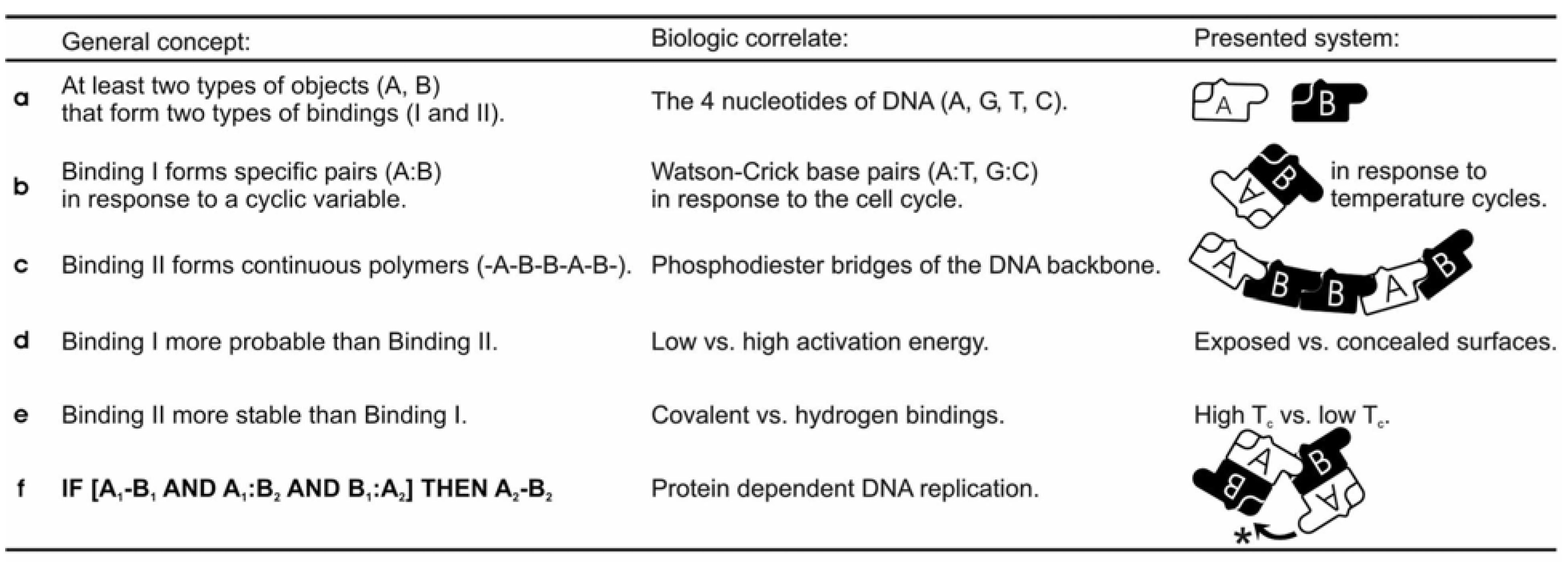
Figure 1.
The general concept of template-replicating aperiodic polymers in relation to DNA and the presented ferromagnetic system. Representation of information as strings demands at least two types of monomeric objects (a), represented by the four nucleotides of DNA, and the A and B constructs of the model system. Template replication is based on two types of bindings occurring between the objects; Binding I forming complementary pairs in response to a cyclic variable (b), and Binding II forming continuous polymers (c). Binding I is more probable than Binding II (d), whereas Binding II is more stabile than Binding I (e), corresponding to the relationship between the covalent and the hydrogen bindings of DNA, and the two different magnetic bindings of the model system (Tc: Curie temperature). The key to formation and transfer of genetic information lies in the steric and energetic relationship between the two bindings, and may be expressed in terms of conditional logic (f). In simple, monomers that bind to a polymer are joined together in a complementary polymer. This mechanism is dependent on protein catalysis in current biological systems, but may be achieved by simple dynamics as demonstrated by the hinging movement between four connected objects of the model system (*).
Figure 1.
The general concept of template-replicating aperiodic polymers in relation to DNA and the presented ferromagnetic system. Representation of information as strings demands at least two types of monomeric objects (a), represented by the four nucleotides of DNA, and the A and B constructs of the model system. Template replication is based on two types of bindings occurring between the objects; Binding I forming complementary pairs in response to a cyclic variable (b), and Binding II forming continuous polymers (c). Binding I is more probable than Binding II (d), whereas Binding II is more stabile than Binding I (e), corresponding to the relationship between the covalent and the hydrogen bindings of DNA, and the two different magnetic bindings of the model system (Tc: Curie temperature). The key to formation and transfer of genetic information lies in the steric and energetic relationship between the two bindings, and may be expressed in terms of conditional logic (f). In simple, monomers that bind to a polymer are joined together in a complementary polymer. This mechanism is dependent on protein catalysis in current biological systems, but may be achieved by simple dynamics as demonstrated by the hinging movement between four connected objects of the model system (*).
![Entropy 03 00273 g001]()
From this algorithmic relationship between “ideal nucleotides”, we concluded that template replication of aperiodic polymeric structures could be achieved from a variety of architectures. To skirt problems related to the manipulation of chemical bounds, and with reference to previous physical models of replication [
19,
20], we decided on a system of macroscopic objects floating freely in liquid. Chaotic interactions, as for molecules in solution, could thereby be achieved by means of liquid dynamics. The objects were designed in AutoCAD2000 (Autodesk Inc., San Rafael, CA, USA), and manufactured from foamed PVC (Vikupor SC, Vink Plast AS, Kolbotn, Norway) with a Micron VCE 750 (Haas Automation Inc., Oxnard, CA, USA).
The major challenge of the study was to make these plastic objects interact by the logic of template replication (
Fig. 1). By embedding permanent neodymium magnets (BM 35, Ø 5 x 2 mm axial, Bakker Magnetics Nordic AS, Stavanger, Norway) and corresponding temporary magnets with Curie temperature (T
c) near the ambient temperature, we came up with magnetic binding forces that fluctuated in response to alterations in temperature, due to phase transition of the soft magnet [
21]. This established an explicit analogy to the thermal effect on chemical bounds, and a direct link between external energy flow and the properties of the individual object. The commercially available alloys, Monel 400 and Monel 405R (HP Alloys Inc., Tipton, IN, USA) were chosen as the temporary magnets of Binding I and Binding II, respectively, with the inactivation of Binding II approximately 30°C above that of Binding I. Two types of objects, A and B, complementary for Binding I and congruent for Binding II (
Fig. 2), were placed in a 100 l thermocycler (custom-built by Pedersen & Sønn AS, Oslo, Norway), and monitored by digital video.
Figure 2.
Three-dimensional representations of the two monomeric objects, A (red) and B (blue). Components of Binding I: Exposed magnets (a) positioned complementary to cylinders of Monel 400 (b), and magnets (c) positioned to block attractive forces between magnets (a) in different objects. Components of Binding II: Partly concealed magnet (d) positioned in correspondence with doom-shaped piece of Monel 405R (e), and a magnet (f) which establishes a resistance in the binding, corresponding to activation energy. Mechanical ridge (g) that inhibited bindings due to magnetic fields spanning through the objects. Steel weight (h) and plastic cap (i). Orientation of magnetic poles are indicated as + and –.
Figure 2.
Three-dimensional representations of the two monomeric objects, A (red) and B (blue). Components of Binding I: Exposed magnets (a) positioned complementary to cylinders of Monel 400 (b), and magnets (c) positioned to block attractive forces between magnets (a) in different objects. Components of Binding II: Partly concealed magnet (d) positioned in correspondence with doom-shaped piece of Monel 405R (e), and a magnet (f) which establishes a resistance in the binding, corresponding to activation energy. Mechanical ridge (g) that inhibited bindings due to magnetic fields spanning through the objects. Steel weight (h) and plastic cap (i). Orientation of magnetic poles are indicated as + and –.
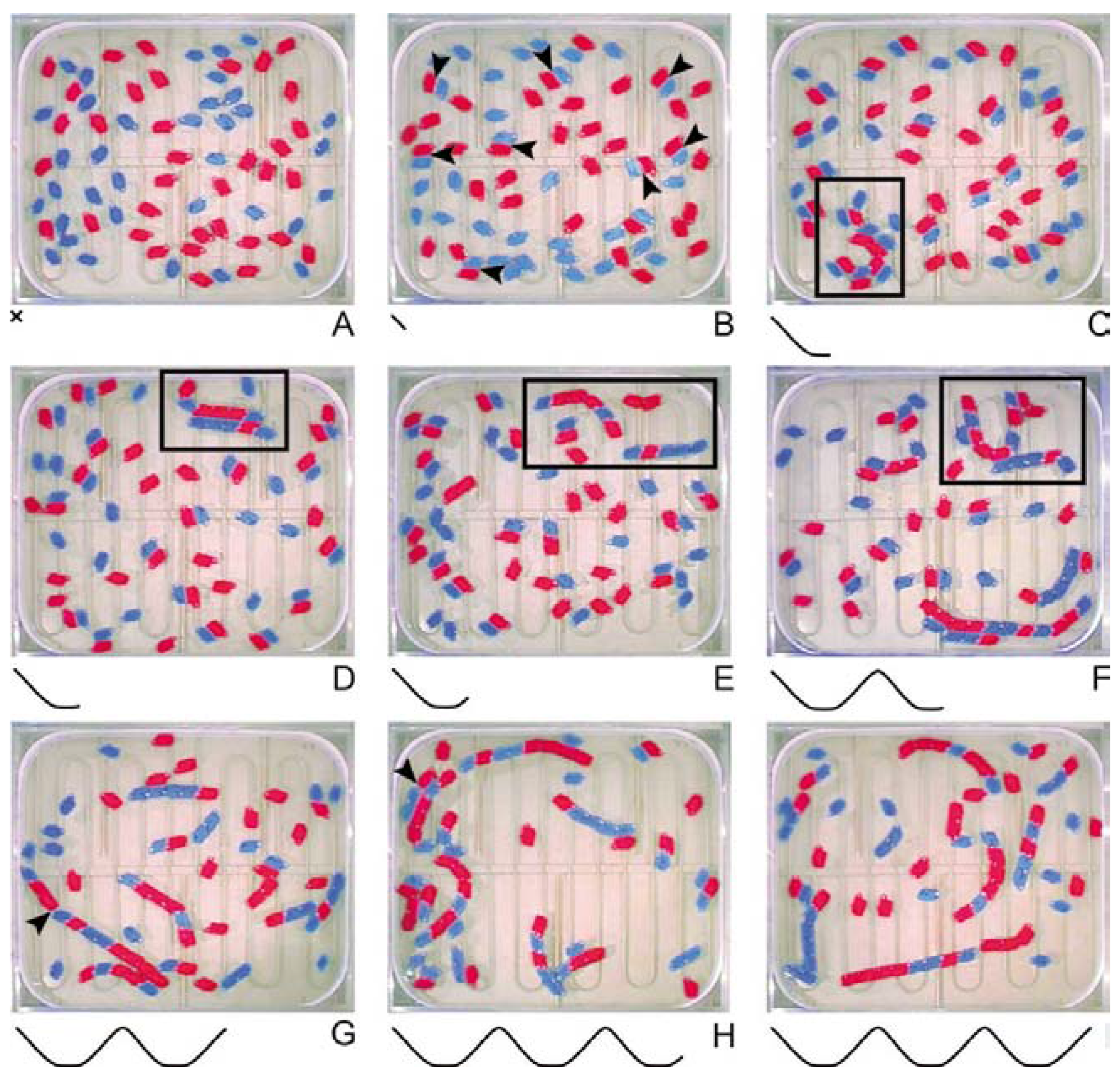
The basic experiment was performed at constant liquid turbulence, and started with 70 objects (35 of each) floating individually at 60°C (
Fig. 3A). Above this temperature there were no stable connections occurring between the objects. The temperature was then lowered to 15°C over a period of 1 hour, and A and B objects started to form stabile pairs through Binding I (
Fig. 3B). Concomitantly, but at a significantly lower rate (approx. 1 to 10) there was spontaneous formation of Binding II (
Fig. 3C). Combinations of the two bindings then resulted in double stranded structures, when objects bound by Binding I to objects bound by Binding II flipped together in Binding II (
Fig. 3C-D; illustrated by the arrow in
Fig. 1). After 30 min at 15°C, the temperature was increased to 60°C; Binding I was broken, and the double stranded structures dissolved into their complementary components (
Fig. 3E). Through repetitive thermocycles, established polymers then acted as templates for the formation of new sequences (
Fig. 3F-I).
Figure 3.
Self-organization of template-replicating polymers through three consecutive thermocycles. Passage through the thermocycles is represented by the extending curve. Key events are indicated. At 60°C all monomeric objects moved independently (A). On lowering the temperature, numerous pair of A (red) and B (blue) objects combined by Binding I (B). A complex comprising a combination of several bindings (I and II) emerged at 15°C (C). The complex flipped into a double-stranded configuration (D). The two strands dissociated after an increase in temperature (E). Dynamic interactions between polymers, monomers and the environment gave rise to secondary and tertiary configurations (F). Long polymers were instable at high temperatures, and breaking of Binding II established an open-ended dynamic (G). A sixmer directed the elongation of a dimer (H), resulting a new generation of polymers (I).
Figure 3.
Self-organization of template-replicating polymers through three consecutive thermocycles. Passage through the thermocycles is represented by the extending curve. Key events are indicated. At 60°C all monomeric objects moved independently (A). On lowering the temperature, numerous pair of A (red) and B (blue) objects combined by Binding I (B). A complex comprising a combination of several bindings (I and II) emerged at 15°C (C). The complex flipped into a double-stranded configuration (D). The two strands dissociated after an increase in temperature (E). Dynamic interactions between polymers, monomers and the environment gave rise to secondary and tertiary configurations (F). Long polymers were instable at high temperatures, and breaking of Binding II established an open-ended dynamic (G). A sixmer directed the elongation of a dimer (H), resulting a new generation of polymers (I).
The polymeric Binding II was designed to be semi-stable at high temperatures (
Fig. 3G) and the combined synthesis and degradation of sequences established an open-ended dynamic. More complex interactions emerged as polymers of different sequences exhibited distinctive behaviors due to the collective magnetomechanical properties of A and B objects (
Fig. 3F). Such properties were observed as sequence-dependent tendencies to form secondary and tertiary configurations, as well as, differences in response to the mechanical boundaries, the static magnetic fields, and the hydrodynamics of the thermocycler. As an example, polymers predominantly composed of A objects had a slight tendency to slide along the perimeter of the thermocycler due to weak attractions between neodymium magnets and the steel walls. These and similar phenotypic characteristics, not anticipated by design, were visually observed to affect the replication of individual sequences.
This model mechanistically demonstrates self-organization of template-replicating aperiodic polymers, and evidently shows that such dynamics are thermodynamically viable. The initial formation of random sequences directs the synthesis of complementary sequences, which logically are not random, and there is an intuitive reduction of uncertainty within the system. Such reduction of uncertainty may be expressed in terms of Shannon’s theory of communication [
22,
23], and relates directly to the definition of information. Reiterated from above, information is defined as shared entropy between two independent structures [
1,
2], and that is exactly the result when complementary polymers split apart. Combined, it may thus be stated that this system demonstrates self-organization of genetic information.
What is this information about? Most importantly, each strand involves a combined program and machinery for building complementary sequences, and related polymers carry information about each others inherent structure. Thereby they also carry information about a common mode of interaction with the surrounding environment, which in turn determines whether a specific sequence is replicated or not. Natural selection, the fundamental concept underlying Darwinian evolution [
7,
8], is thus demonstrated in an intuitively evident manner, and it may be argued that the evolving polymers carry information, not only about their line of related structures, but also about the environment that allowed them to propagate [
8,
9,
24]. To statistically confirm Darwinian evolution of increasing fitness within such a physical system will require more extensive experiments with a larger number of monomers.
The sequential information is by no means programmed into the design of the system, but self-organizes from chaotic interactions exclusively driven by fluctuations in heat and turbulence. Interestingly, a similar relationship between thermodynamics and information has for more than a century been illustrated by the conundrum of the Maxwell Demon [
25]. This hypothetical being uses its information-processing ability to reduce the entropy of a gas [
4], and several authors have drawn attention to similarities between the Maxwell Demon and the fundamental dynamics of living systems [
10,
26,
27]. Adami
et al. have demonstrated this relationship by analyzing accumulation of complexity in a computer based evolution system [
9], but a physical mechanism that unites thermodynamics and information theory in an evident manner has never been presented.
This study shows that such a mechanism can be directly derived from the fundamentals of molecular biology, and demonstrates spontaneous rise of genetic information from chaotic interactions between physical objects. In essence, information is produced by the flow of energy passing through the thermocycler, and the system thereby visually and mechanistically demonstrates that there indeed is a direct link between the thermodynamic and the information theoretical concept of entropy [
1,
3,
4]. How these dynamics translate into mathematics is beyond the scope of this study, but the model offers a useful perspective for exploring this fundamental question.
Although this is a non-chemical and very simplistic model we believe that the study has important implications for the understanding of living systems. The model visually and mechanistically demonstrates a fundamental relationship between thermodynamics, information theory, and life science that can be very difficult to grasp from mathematical or semantic abstractions [
17]. It has immediate applications as a tool for simulating and communicating basic concepts related to a broad field of science, and vividly visualizes the basic molecular concept underlying the biotech revolution. Moreover, this model is to our knowledge the first demonstration of a physical contraption with a potential for Darwinian evolution. It has recently been argued that such systems will open for a new generation of technologies [
28,
29], and the prospect of applying this simple algorithmic concept (
Fig. 1) to nano-particles, mobile robotics, or even virtual constructs on the Internet is highly interesting.
In conclusion, this model didactically demonstrates the fundamental connection between physics and life science, and may involve an important contribution in the ongoing quest to develop living and intelligent technology.