A New Method for the Determination of Nucleic Acid Using an Eu3+– nicotinic Acid Complex as a Resonance Light Scattering Probe
Abstract
:Introduction
Results and Discussion
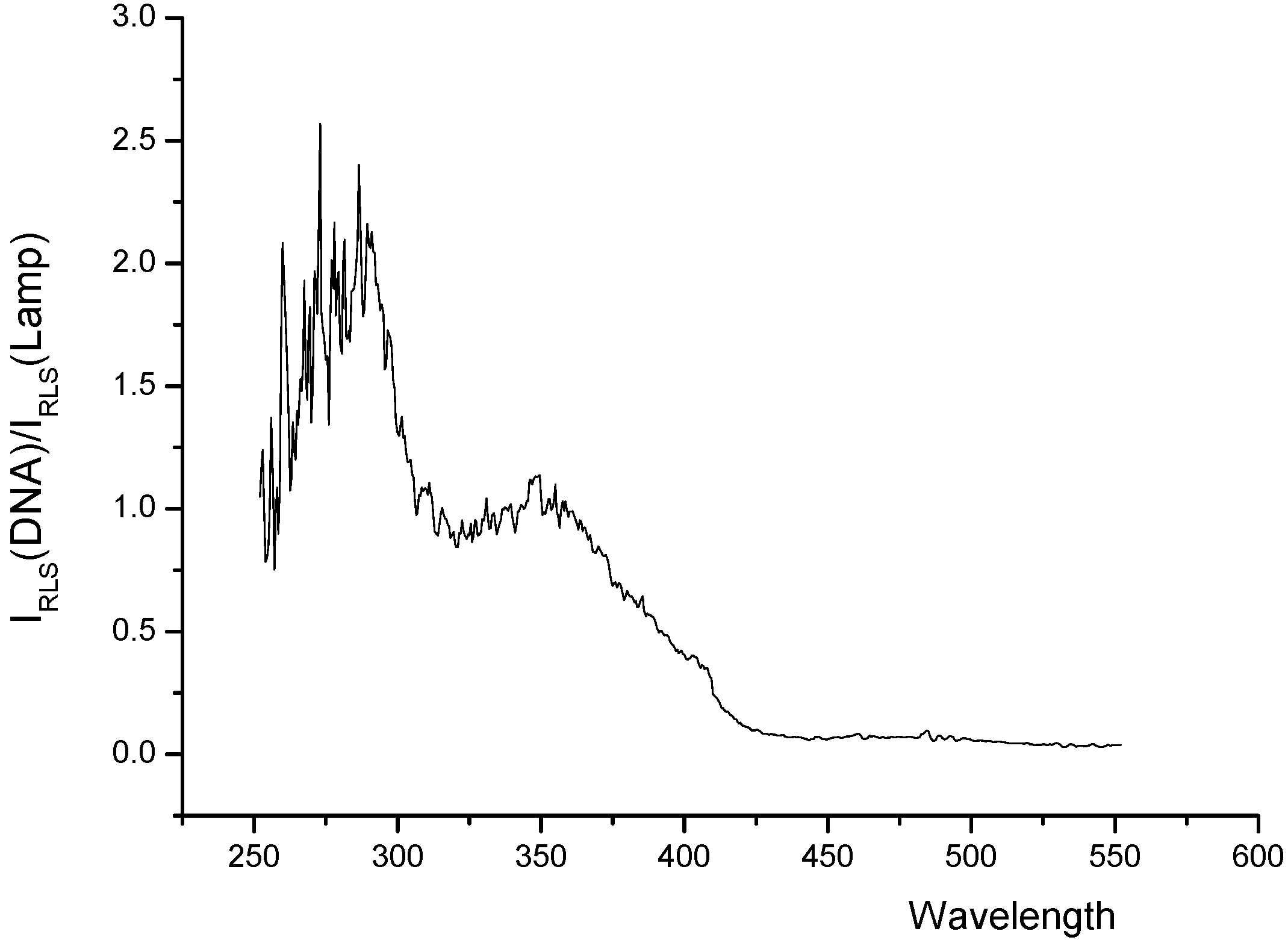
RLS spectra of the systems

Optimization of conditions for RLS measurements

| Buffers | Tris-HCl | HMTA*-HCl | NaH2PO4-K2HPO4 | NH4Cl-NH3 | NaH2PO4-Citric acid |
|---|---|---|---|---|---|
| ΔIR(%) | 100 | 50.9 | 12.8 | 45.9 | 49.3 |
| M3+ | Eu3+ | Tb3+ | Y3+ | Nd3+ | Al3+ | Dy3+ | Gd3+ |
|---|---|---|---|---|---|---|---|
| IR(%) | 100 | 46.4 | 99.0 | 54.6 | 66.6 | 68.2 | 65.0 |


Analytical application
| Probe | Nucleic acid | Detection limit (10-9g∙mL-1) | References |
|---|---|---|---|
| TAPP*1 | ctDNA/fsDNA/yRNA | 4.1/4.6/6.7 | [4] |
| Safranine T | ctDNA/fsDNA/yRNA | 13.2/39.8/61.1 | [5] |
| Neutral Red | ctDNA/fsDNA/yRNA | 48.2/35.2/205 | [6] |
| BCB*2 | ctDNA/fsDNA/yRNA | 118/112/434 | [7] |
| Methyl Green | ctDNA/fsDNA/yRNA | 7.8/2.6/9.9 | [9] |
| Azur A | ctDNA/fsDNA | 19.9/12.6 | [11] |
| Crystal Violet | ctDNA/fsDNA/yRNA | 13.8/36.8/69 | [8] |
| Methyl Violet | DNA | 100 | [10] |
| Congo Red | fsDNA/ctDNA/yRNA | 0.019/ 0.89/1.2 | [12] |
| Methyl green | fsDNA | 2.6/7.8/9.9 | [13] |
| OA*3–Eu3+ | fsDNA/ctDNA/yRNA | 0.02/0.011/0.01 | [14] |
| Eu3+-TTA-Phen | fsDNA/yRNA/ctDNA | 0.03/0.006/0.002 | [15] |

Conclusions
Experimental
General
Acknowledgements
References and Notes
- Udenfriend, S.; Zaltzman, P. Fluorescence characteristics of purines pyrimines and their derives. Anal. Biochem. 1962, 3, 49–59. [Google Scholar] [CrossRef]
- Wang, Y.B.; Yang, J.H.; Wu, X.; Li, L.; Sun, S.N.; Su, B.Y. Progress of the Spectral Probe for Nucleic Acids. Anal. Lett. 2003, 36, 2063–2094. [Google Scholar] [CrossRef]
- Pasternack, R.F.; Bustamante, C.; Collings, P.J.; Giannetteo, A.; Gibbs, E.J. Porphyrin assemblies on DNA as studied by a resonance light-scattering technique. J. Am. Chem. Soc. 1993, 115, 5393–5399. [Google Scholar] [CrossRef]
- Huang, C.Z.; Li, K.A.; Tong, S.Y. Determination of nucleic acids by a resonance light-scattering technique with a,b,g,d-tetrakis [4-(trimethylammoniumyl) phenyl] porphine. Anal. Chem. 1996, 68, 2259–2263. [Google Scholar] [CrossRef]
- Huang, C.Z.; Li, Y.F.; Liu, X.D. Determination of nucleic acids at nanogram levels with safranine T by a resonance light-scattering technique. Anal. Chim. Acta 1998, 375, 89–97. [Google Scholar] [CrossRef]
- Wang, Y.T.; Zhao, F.L.; Li, K.A.; Tong, S.Y. Molecular spectroscopic study of DNA binding with neutral red and application to assay of nucleic acids. Anal. Chim. Acta 1999, 396, 75–81. [Google Scholar] [CrossRef]
- Wang, Y.T.; Zhao, F.L.; Li, K.A.; Tong, S.Y. Molecular spectroscopy study of the reaction of nucleic acids with brilliant cresol blue. Spectrochim. Acta. Part A. 2000, 56, 1827–1833. [Google Scholar] [CrossRef]
- Zhang, W.J.; Xu, H.P.; Wu, S.Q.; Chen, X.G.; Hu, Z.D. Determination of nucleic acids with crystal violet. Analyst 2001, 126, 513–517. [Google Scholar] [CrossRef]
- Liu, R.T.; Yang, J.H.; Wu, X.; Sun, C.X. Simple and sensitive determination of nucleic acid by Rayleigh light scattering technique with methyl green-CTMAB. Indian J. Chem. 2001, 40A, 1121–1123. [Google Scholar]
- Liu, Y.; Ma, C.Q.; Li, K.A.; Xie, F.C.; Tong, S.Y. Rayleigh light scattering study on the reaction of nucleic acids and methyl violet. Anal. Biochem. 1999, 268, 187–192. [Google Scholar] [CrossRef]
- Li, Y.F.; Huang, C.Z.; Huang, X.H.; Li, M. Determination of DNA by its enhancement effect of resonance light scattering by azur A. Anal. Chim. Acta 2001, 429, 311–319. [Google Scholar] [CrossRef]
- Wu, X.; Wang, Y.B.; Yang, J.H.; Sun, S.N. Determination of nucleic acids at nanogram level using resonance light scattering technique with Congo Red. Spectrochim. Acta 2005, 61A, 361–366. [Google Scholar]
- Liu, R.T.; Yang, J.H.; Wu, X.; Sun, C.X. Simple and sensitive determination of nucleic acid by Rayleigh light scattering technique with methyl green-CTMAB. Indian J. Chem. 2001, 40A, 1121–1123. [Google Scholar]
- Wu, X.; Sun, S.N.; Yang, J.H.; Wang, M,Q.; Liu, L.Y.; Guo, C.Y. Study on the interaction between nucleic acid and Eu3+-oxolinic acid and the determination of nucleic acid using the Resonance Light Scattering technique. Spectrochim. Acta 2005, 62A, 896–901. [Google Scholar]
- Jia, Z.; Yang, J.H.; Wu, X.; Sun, C.X.; Liu, S.F.; Wang, F. The sensitive determination of nucleic acids using resonance light scattering quenching method. Spectrochim. Acta Part A. 2006, 64A, 555–559. [Google Scholar]
- Collins, F.S.; Patrinos, A.; Jordan, E.; Chakravi, A.; Gesteland, R.; Walters, L. New goals for the U.S. Human Genome Project. Science 1998, 282, 682. [Google Scholar]
- McPhie, P. Resonance light scattering profiles must be corrected for instrument performance. Anal. Biochem. 2006, 348, 157–159. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the synthetic and cucumber are available from authors.
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Guo, M.; Wang, L.-T.; Wu, X.; Xu, W.; Yang, J.-H. A New Method for the Determination of Nucleic Acid Using an Eu3+– nicotinic Acid Complex as a Resonance Light Scattering Probe. Molecules 2009, 14, 10-18. https://doi.org/10.3390/molecules14010010
Guo M, Wang L-T, Wu X, Xu W, Yang J-H. A New Method for the Determination of Nucleic Acid Using an Eu3+– nicotinic Acid Complex as a Resonance Light Scattering Probe. Molecules. 2009; 14(1):10-18. https://doi.org/10.3390/molecules14010010
Chicago/Turabian StyleGuo, Meng, Lin-Tong Wang, Xia Wu, Wei Xu, and Jing-He Yang. 2009. "A New Method for the Determination of Nucleic Acid Using an Eu3+– nicotinic Acid Complex as a Resonance Light Scattering Probe" Molecules 14, no. 1: 10-18. https://doi.org/10.3390/molecules14010010
APA StyleGuo, M., Wang, L.-T., Wu, X., Xu, W., & Yang, J.-H. (2009). A New Method for the Determination of Nucleic Acid Using an Eu3+– nicotinic Acid Complex as a Resonance Light Scattering Probe. Molecules, 14(1), 10-18. https://doi.org/10.3390/molecules14010010




