Study on Suitability of KOD DNA Polymerase for Enzymatic Production of Artificial Nucleic Acids Using Base/Sugar Modified Nucleoside Triphosphates
Abstract
:1. Introduction
2. Results and Discussion
3. Experimental
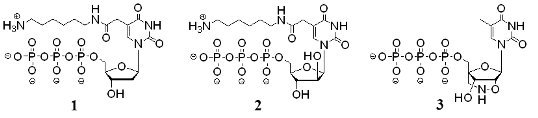
3.1. Synthesis of modified nucleoside triphosphate analogs and their intermediates
3.2. Preparation of mutated KOD DNA polymerases
3.3. Single nucleotide incorporation in primer extension reaction using KOD mutants and Vent(exo-) DNA polymerase
3.4. Successive incorporation of 2′,4′-bridged nucleotides using analogue 3 with various DNA polymerases
4. Conclusions
Acknowledgements
References and Notes
- Robertson, D.L.; Joyce, G.F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 1990, 344, 467–468. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.E.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Kuwahara, M.; Sugimoto, N. Molecular evolution of functional nucleic acids with chemical modifications. Molecules 2010, 15, 5423–5444. [Google Scholar] [CrossRef] [PubMed]
- Kempeneers, V.; Renders, M.; Froeyen, M.; Herdewijn, P. Investigation of the DNA-dependent cyclohexenyl nucleic acid polymerization and the cyclohexenyl nucleic acid-dependent DNA polymerization. Nucleic Acids Res. 2005, 33, 3828–3836. [Google Scholar] [CrossRef] [PubMed]
- Horhota, A.; Zou, K.; Ichida, J.K.; Yu, B.; McLaughlin, L.W.; Szostak, J.W.; Chaput, J.C. Kinetic analysis of an efficient DNA-dependent TNA polymerase. J. Am. Chem. Soc. 2005, 127, 7427–7434. [Google Scholar] [CrossRef] [PubMed]
- Leal, N.A.; Sukeda, M.; Benner, S.A. Dynamic assembly of primers on nucleic acid templates. Nucleic Acids Res. 2006, 34, 4702–4710. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.H.; Chen, J.; Szostak, J.W. Enzymatic synthesis of DNA on glycerol nucleic acid templates without stable duplex formation between product and template. Proc. Natl. Acad. Sci. USA 2007, 104, 14598–14603. [Google Scholar] [CrossRef] [PubMed]
- Lam, C.; Hipolito, C.; Perrin, D.M. Synthesis and enzymatic incorporation of modified deoxyadenosine triphosphates. Eur. J. Org. Chem. 2008, 29, 4915–4923. [Google Scholar] [CrossRef]
- Kuwahara, M.; Takeshima, H.; Nagashima, J.; Minezaki, S.; Ozaki, H.; Sawai, H. Transcription and reverse transcription of artificial nucleic acids involving backbone modification by template-directed DNA polymerase reactions. Bioorg. Med. Chem. 2009, 17, 3782–3788. [Google Scholar] [CrossRef] [PubMed]
- El-Sagheer, A.H.; Brown, T. Synthesis and polymerase chain reaction amplification of DNA strands containing an unnatural triazole linkage. J. Am. Chem. Soc. 2009, 131, 3958–3964. [Google Scholar] [CrossRef] [PubMed]
- Weisbrod, S.H.; Marx, A. Novel strategies for the site-specific covalent labelling of nucleic acids. Chem. Commun. 2008, 44, 5675–5685. [Google Scholar] [CrossRef] [PubMed]
- Hocek, M.; Fojta, M. Cross-coupling reactions of nucleoside triphosphates followed by polymerase incorporation. Construction and applications of base-functionalized nucleic acids. Org. Biomol. Chem. 2008, 6, 2233–2241. [Google Scholar] [CrossRef] [PubMed]
- Loakes, D.; Holliger, P. Polymerase engineering: towards the encoded synthesis of unnatural biopolymers. Chem. Commun. 2009, 31, 4619–4631. [Google Scholar] [CrossRef] [PubMed]
- Ikonen, S.; Macícková-Cahová, H.; Pohl, R.; Sanda, M.; Hocek, M. Synthesis of nucleoside and nucleotide conjugates of bile acids, and polymerase construction of bile acid-functionalized DNA. Org. Biomol. Chem. 2010, 8, 1194–1201. [Google Scholar] [CrossRef] [PubMed]
- Sawai, H.; Ozaki, A.N.; Satoh, F.; Ohbayashi, T.; Masud, M.M.; Ozaki, H. Expansion of structural and functional diversities of DNA using new 5-substituted deoxyuridine derivatives by PCR with superthermophilic KOD Dash DNA polymerase. Chem. Commun. 2001, 24, 2604–2605. [Google Scholar] [CrossRef]
- Obayashi, T.; Masud, M.M.; Ozaki A., N.; Ozaki, H.; Kuwahara, M.; Sawai, H. Enzymatic synthesis of labeled DNA by PCR using new fluorescent thymidine nucleotide analogue and superthermophilic KOD dash DNA polymerase. Bioorg. Med. Chem. Lett. 2002, 12, 1167–1170. [Google Scholar] [CrossRef]
- Ohbayashi, T.; Kuwahara, M.; Hasegawa, M.; Kasamatsu, T.; Tamura, T.; Sawai, H. Expansion of repertoire of modified DNAs prepared by PCR using KOD Dash DNA polymerase. Org. Biomol. Chem. 2005, 3, 2463–2468. [Google Scholar] [CrossRef] [PubMed]
- Kuwahara, M.; Hanawa, K.; Ohsawa, K.; Kitagata, R.; Ozaki, H.; Sawai, H. Direct PCR amplification of various modified DNAs having amino acids: convenient preparation of DNA libraries with high-potential activities for in vitro selection. Bioorg. Med. Chem. 2006, 14, 2518–2526. [Google Scholar] [CrossRef] [PubMed]
- Sawai, H.; Nagashima, J.; Kuwahara, M.; Kitagata, R.; Tamura, T.; Matsui, I. Differences in substrate specificity of C(5)-substituted or C(5)-unsubstituted pyrimidine nucleotides by DNA polymerases from thermophilic bacteria, archaea, and phages. Chem. Biodivers. 2007, 4, 1979–1995. [Google Scholar] [CrossRef] [PubMed]
- Veedu, R.N.; Vester, B.; Wengel, J. Efficient enzymatic synthesis of LNA-modified DNA duplexes using KOD DNA polymerase. Org. Biomol. Chem. 2009, 7, 1404–1409. [Google Scholar] [CrossRef] [PubMed]
- Veedu, R.N.; Wengel, J. Locked nucleic acid nucleoside triphosphates and polymerases: on the way towards evolution of LNA aptamers. Mol. Biosyst. 2009, 5, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Obika, S.; Nanbu, D.; Hari, Y.; Morio, K.; In, Y.; Ishida, T.; Imanishi, T. Synthesis of 2′-O,4′-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C3′-endo sugar puckering. Tetrahedron Lett. 1997, 38, 8735–8738. [Google Scholar] [CrossRef]
- Obika, S.; Nanbu, D.; Hari, Y.; Andoh, J.; Morio, K.; Doi, T.; Imanishi, T. Stability and structural features of the duplexes containing nucleoside analogs with a fixed N-type conformation, 2′-O,4′-C-methyleneribonucleosides. Tetrahedron Lett. 1998, 39, 5401–5404. [Google Scholar] [CrossRef]
- Singh, S.K.; Nielsen, P.; Koshkin, A.A.; Wengel, J. LNA (locked nucleic acids): synthesis and high-affinity nucleic acid recognition. Chem. Commun. 1998, 4, 455–456. [Google Scholar] [CrossRef]
- Wirges, C.T.; Timper, J.; Fischler, M.; Sologubenko, A.S.; Mayer, J.; Simon, U.; Carell, T. Controlled nucleation of DNA metallization. Angew. Chem. Int. Ed. 2009, 48, 219–223. [Google Scholar] [CrossRef] [PubMed]
- Raindlová, V.; Pohl, R.; Sanda, M.; Hocek, M. Direct polymerase synthesis of reactive aldehyde-functionalized DNA and its conjugation and staining with hydrazines. Angew. Chem. Int. Ed. 2010, 49, 1064–1066. [Google Scholar] [CrossRef] [PubMed]
- Takagi, M.; Nishioka, M.; Kakihara, H.; Kitabayashi, M.; Inoue, H.; Kawakami, B.; Oka, M.; Imanaka, T. Characterization of DNA polymerase from Pyrococcus sp. strain KOD1 and its application to PCR. Appl. Environ. Microbiol. 1997, 63, 4504–4510. [Google Scholar] [PubMed]
- Nishioka, M.; Mizuguchi, H.; Fujiwara, S.; Komatsubara, S.; Kitabayashi, M.; Uemura, H.; Takagi, M.; Imanaka, T. Long and accurate PCR with a mixture of KOD DNA polymerase and its exonuclease deficient mutant enzyme. J. Biotechnol. 2001, 88, 141–149. [Google Scholar] [CrossRef]
- Kuroita, T.; Matsumura, H.; Yokota, N.; Kitabayashi, M.; Hashimoto, H.; Inoue, T.; Imanaka, T.; Kai, Y. Structural mechanism for coordination of proofreading and polymerase activities in Archaeal DNA polymerases. J. Mol. Biol. 2005, 351, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Perler, F.B.; Comb, D.G.; Jack, W.E.; Moran, L.S.; Qiang, B.; Kucera, R.B.; Benner, J.; Slatko, B.E.; Nwankwo, D.O.; Hempstead, S.K.; Carlow, C.K.S.; Jannasch, H. Intervening sequences in an Archaea DNA polymerase gene. Proc. Natl. Acad. Sci. USA 1992, 89, 5577–81. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Kucera, R.B.; Jack, W.E. Characterization of a DNA polymerase from the hyperthermophile archaea Thermococcus litoralis. Vent DNA polymerase, steady state kinetics, thermal stability, processivity, strand displacement, and exonuclease activities. J. Biol. Chem. 1993, 268, 1965–1975. [Google Scholar] [PubMed]
- Kuwahara, M.; Nagashima, J.; Hasegawa, M.; Tamura, T.; Kitagata, R.; Hanawa, K.; Hososhima, S.; Kasamatsu, T.; Ozaki, H.; Sawai, H. Systematic characterization of 2′-deoxynucleoside- 5′-triphosphate analogs as substrates for DNA polymerases by polymerase chain reaction and kinetic studies on enzymatic production of modified DNA. Nucleic Acids Res. 2006, 34, 5383–5394. [Google Scholar] [CrossRef] [PubMed]
- Lanford, R.E.; Hildebrandt-Eriksen E., S.; Petri, A.; Persson, R.; Lindow, M.; Munk, M.E.; Kauppinen, S.; Ørum, H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 2010, 327, 198–201. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Fisker, N.; Asselin, M.C.; Lindholm, M.; Rosenbohm, C.; Ørum, H.; Elmén, J.; Seidah, N.G.; Straarup, E.M. A locked nucleic acid antisense oligonucleotide (LNA) silences PCSK9 and enhances LDLR expression in vitro and in vivo. PLoS One 2010, 5, e10682. [Google Scholar] [CrossRef] [PubMed]
- Kuwahara, M.; Obika, S.; Nagashima, J.; Ohta, Y.; Suto, Y.; Ozaki, H.; Sawai, H.; Imanishi, T. Systematic analysis of enzymatic DNA polymerization using oligo-DNA templates and triphosphate analogs involving 2′,4′-bridged nucleosides. Nucleic Acids Res. 2008, 36, 4257–4265. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.M.; Seki, S.; Obika, S.; Haitani, S.; Miyashita, K.; Imanishi, T. Highly stable pyrimidine-motif triplex formation at physiological pH values by a bridged nucleic acid analog. Angew. Chem. Int. Ed. 2007, 46, 4306–4309. [Google Scholar] [CrossRef] [PubMed]
- Kuwahara, M.; Obika, S.; Takeshima, H.; Hagiwara, Y.; Nagashima, J.; Ozaki, H.; Sawai, H.; Imanishi, T. Smart conferring of nuclease resistance to DNA by 3′-end protection using 2′,4′-bridged nucleoside-5′-triphosphates. Bioorg. Med. Chem. Lett. 2009, 36, 2941–2943. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, Y.; Kitadume, S.; Morihiro, K.; Kuwahara, M.; Ozaki, H.; Sawai, H.; Imanishi, T.; Obika, S. Effect of 3′-end capping of aptamer with various 2′,4′-bridged nucleotides: enzymatic post-modification toward a practical use of polyclonal aptamers. Bioorg. Med. Chem. Lett. 2010, 20, 1626–1629. [Google Scholar] [CrossRef] [PubMed]
- Creighton, S.; Bloom, L.B.; Goodman, M.F. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 1995, 262, 232–256. [Google Scholar] [PubMed]
- Rahman, S.M.; Seki, S.; Obika, S.; Yoshikawa, H.; Miyashita, K.; Imanishi, T. Design, synthesis, and properties of 2′,4′-BNANC: a bridged nucleic acid analogue. J. Am. Chem. Soc. 2008, 130, 4886–4896. [Google Scholar] [CrossRef] [PubMed]
- Jäger, S.; Rasched, G.; Kornreich-Leshem, H.; Engeser, M.; Thum, O.; Famulok, M. A versatile toolbox for variable DNA functionalization at high density. J. Am. Chem. Soc. 2005, 127, 15071–15082. [Google Scholar] [CrossRef] [PubMed]
- Veedu, R.N.; Vester, B.; Wengel, J. Enzymatic incorporation of LNA nucleotides into DNA strands. ChemBioChem 2007, 8, 490–492. [Google Scholar] [CrossRef] [PubMed]
- Sawai, H.; Nakamura, A.; Sekiguchi, S.; Yumoto, K.; Endoh, M.; Ozaki, H. Efficient synthesis of new 5-substituted uracil nucleosides useful for linker arm incorporation. J. Chem. Soc., Chem. Comm. 1994, 1997–1998. [Google Scholar] [CrossRef]
- Mendelman, L.V.; Boosalis, M.S.; Petruska, J.; Goodman, M.F. Nearest neighbor influences on DNA polymerase insertion fidelity. J. Biol. Chem. 1989, 264, 14415–14423. [Google Scholar] [PubMed]
Sample Availability: The new analog, modified nucleoside triphosphate 2 is available free to academic researchers under a Material Transfer Agreement (MTA). |



| DNA polymerase | dNTP | Vmax (min–1) | Km (mM) | Insertion efficiency Vmax/Km | Accuracyb |
|---|---|---|---|---|---|
| KOD1 | T | 1.8 | 0.021 | 88 | 1 |
| 1 | 11 | 0.14 | 81 | 0.92 | |
| 2 | 0.10 | 0.26 | 3.8 | 0.043 | |
| C | 0.074 | 0.66 | 0.11 | 0.0013 | |
| KOD2 | T | 2.1 | 0.035 | 60 | 1 |
| 1 | 4.0 | 0.083 | 48 | 0.80 | |
| 2 | 0.32 | 0.099 | 3.3 | 0.055 | |
| C | 0.019 | 0.11 | 0.17 | 0.0029 | |
| KOD3 | T | 2.3 | 0.047 | 49 | 1 |
| 1 | 4.4 | 0.081 | 55 | 1.1 | |
| 2 | 0.27 | 0.099 | 2.7 | 0.055 | |
| C | 0.033 | 0.70 | 0.048 | 0.00097 | |
| KOD8 | T | 9.5 | 0.063 | 150 | 1 |
| 1 | 53 | 0.31 | 170 | 1.1 | |
| 2 | 2.4 | 0.38 | 6.3 | 0.041 | |
| C | 0.11 | 1.0 | 0.11 | 0.00073 | |
| Vent(exo-) | T | 0.34 | 0.0030 | 110 | 1 |
| 1 | 0.29 | 0.0027 | 110 | 0.95 | |
| 2 | 0.049 | 0.0044 | 11 | 0.099 | |
| C | 0.14 | 1.7 | 0.083 | 0.00073 |
© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kuwahara, M.; Takano, Y.; Kasahara, Y.; Nara, H.; Ozaki, H.; Sawai, H.; Sugiyama, A.; Obika, S. Study on Suitability of KOD DNA Polymerase for Enzymatic Production of Artificial Nucleic Acids Using Base/Sugar Modified Nucleoside Triphosphates. Molecules 2010, 15, 8229-8240. https://doi.org/10.3390/molecules15118229
Kuwahara M, Takano Y, Kasahara Y, Nara H, Ozaki H, Sawai H, Sugiyama A, Obika S. Study on Suitability of KOD DNA Polymerase for Enzymatic Production of Artificial Nucleic Acids Using Base/Sugar Modified Nucleoside Triphosphates. Molecules. 2010; 15(11):8229-8240. https://doi.org/10.3390/molecules15118229
Chicago/Turabian StyleKuwahara, Masayasu, Yuuki Takano, Yuuya Kasahara, Hiroki Nara, Hiroaki Ozaki, Hiroaki Sawai, Akio Sugiyama, and Satoshi Obika. 2010. "Study on Suitability of KOD DNA Polymerase for Enzymatic Production of Artificial Nucleic Acids Using Base/Sugar Modified Nucleoside Triphosphates" Molecules 15, no. 11: 8229-8240. https://doi.org/10.3390/molecules15118229