A Therapeutic Approach to Nasopharyngeal Carcinomas by DNAzymes Targeting EBV LMP-1 Gene
Abstract
:1. Introduction
2. Results and Discussion
2.1. Design and Chemical Modifications of LMP1 Targeted DNAzymes

2.2. Down-regulation of LMP1 Expression in NPC Cells
2.3. Impact of Dz1 on Major Signal Pathways in NPC Cells
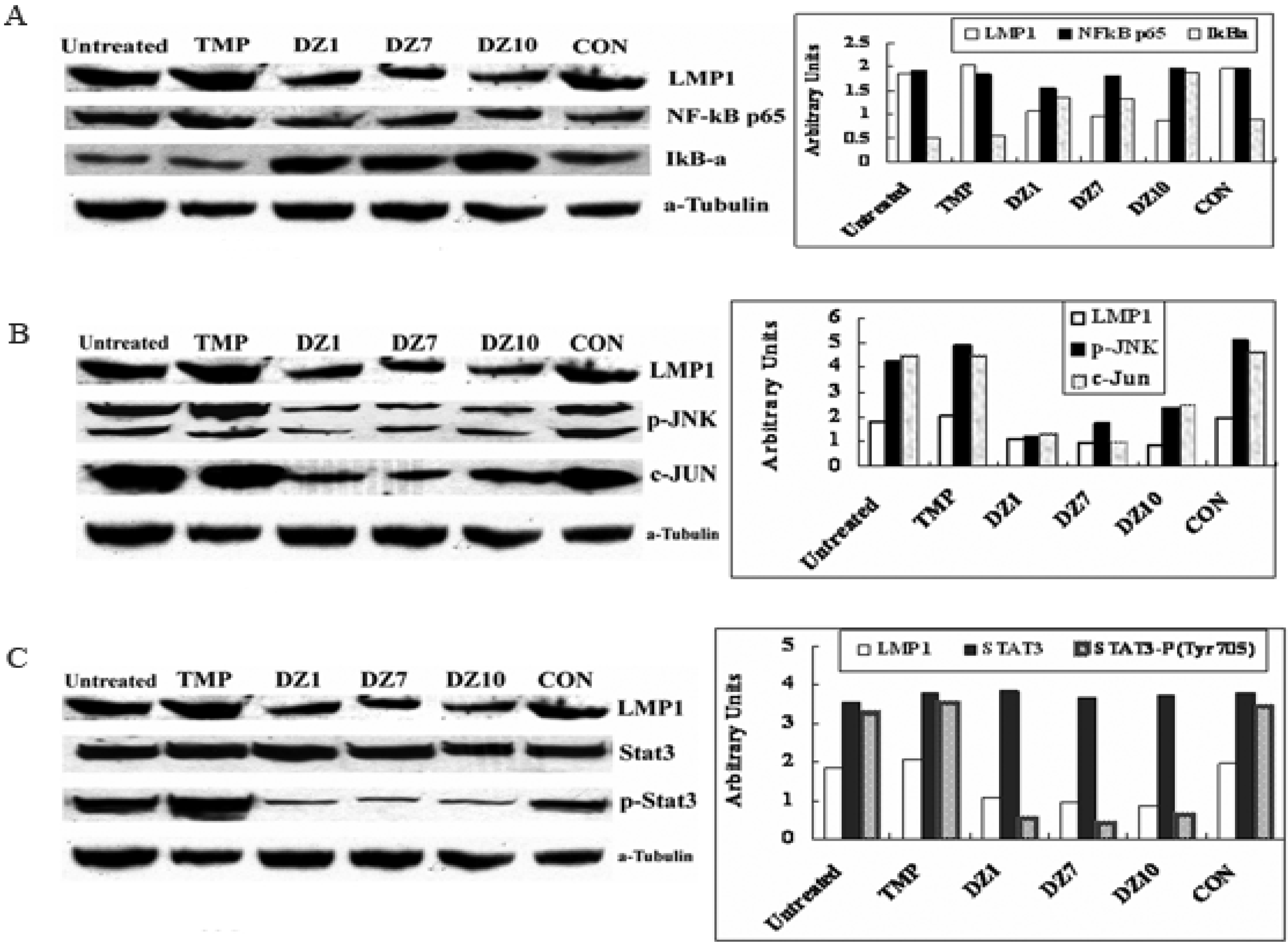


2.4. Biological Effects of DZ1 on NPC
2.4.1. Proliferation
2.4.2. Apoptosis

2.4.3. Metastasis

2.4.4. Anti-tumor effect
2.4.5. Radiosensitization

3. Conclusions

Acknowledgements
References
- Yu, M.C.; Yuan, J.M. Epidemiology of nasopharyngeal carcinoma. Semin. Cancer Biol. 2002, 12, 421–429. [Google Scholar] [CrossRef]
- Herrmann, K.; Niedobitek, G. Epstein-Barr virus-associated carcinomas: Facts and fiction. J. Pathol. 2003, 199, 140–145. [Google Scholar] [CrossRef]
- Chou, J.; Lin, Y.C.; Kim, J.; You, L.; Xu, Z.; He, B.; Jablons, D.M. Nasopharyngeal carcinoma--review of the molecular mechanisms of tumorigenesis. Head Neck 2008, 30, 946–963. [Google Scholar] [CrossRef]
- Tsao, S.W.; Tramoutanis, G.; Dawson, C.W.; Lo, A.K.; Huang, D.P. The significance of LMP1 expression in nasopharyngeal carcinoma. Semin. Cancer Biol. 2002, 12, 473–487. [Google Scholar] [CrossRef]
- Morris, M.A.; Dawson, C.W.; Young, L.S. Role of the Epstein-Barr virus-encoded latent membrane protein-1, LMP1, in the pathogenesis of nasopharyngeal carcinoma. Future Oncol. 2009, 5, 811–825. [Google Scholar] [CrossRef]
- Eliopoulos, A.G.; Young, L.S. LMP1 structure and signal transduction. Semin. Cancer. Biol. 2001, 11, 435–444. [Google Scholar] [CrossRef]
- Zheng, H.; Li, L.L.; Hu, D.S.; Deng, X.Y.; Cao, Y. Role of Epstein-Barr virus encoded latent membrane protein 1 in the carcinogenesis of nasopharyngeal carcinoma. Cell Mol. Immunol. 2007, 4, 185–196. [Google Scholar]
- Hatzivassiliou, E.; Mosialos, G. Cellular signaling pathways engaged by the Epstein-Barr virus transforming protein LMP1. Front Biosci. 2002, 7, d319–d329. [Google Scholar] [CrossRef]
- Li, H.P.; Chang, Y.S. Epstein-Barr virus latent membrane protein 1: Structure and functions. J. Biomed. Sci. 2003, 10, 490–504. [Google Scholar] [CrossRef]
- Horikawa, T.; Yang, J.; Kondo, S.; Yoshizaki, T.; Joab, I.; Furukawa, M.; Pagano, J.S. Twist and epithelial-mesenchymal transition are induced by the EBV oncoprotein latent membrane protein 1 and are associated with metastatic nasopharyngeal carcinoma. Cancer Res. 2007, 67, 1970–1978. [Google Scholar] [CrossRef]
- Dawson, C.W.; Laverick, L.; Morris, M.A.; Tramoutanis, G.; Young, L.S. Epstein-Barr virus-encoded LMP-1 regulates epithelial cell motility and invasion via the Erk-MAPK pathway. J. Virol. 2008, 82, 3654–3664. [Google Scholar] [CrossRef]
- Breaker, R.R.; Joyce, G.F. A DNA enzyme that cleaves RNA. Chem. Biol. 1994, 1, 223–229. [Google Scholar] [CrossRef]
- Breaker, R.R.; Joyce, G.F. A DNA enzyme with Mg(2+)-dependent RNA phosphoesterase activity. Chem. Biol. 1995, 2, 655–660. [Google Scholar] [CrossRef]
- Santoro, S.W.; Joyce, G.F. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. USA 1997, 94, 4262–4266. [Google Scholar] [CrossRef]
- Santoro, S.W.; Joyce, G.F. Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry 1998, 37, 13330–13342. [Google Scholar] [CrossRef]
- Breaker, R.R. DNA enzymes. Nat. Biotechnol. 1997, 15, 427–431. [Google Scholar] [CrossRef]
- Li, Y.; Breaker, R.R. Deoxyribozymes: New players in the ancient game of biocatalysis. Curr. Opin. Struct. Biol. 1999, 9, 315–323. [Google Scholar] [CrossRef]
- Dass, C.R.; Choong, P.F.; Khachigian, L.M. DNAzyme technology and cancer therapy: Cleave and let die. Mol. Cancer Ther. 2008, 7, 243–251. [Google Scholar]
- Willner, I.; Shlyahovsky, B.; Zayats, M.; Willner, B. DNAzymes for sensing, nanobiotechnology and logic gate applications. Chem. Soc. Rev. 2008, 37, 1153–1165. [Google Scholar] [CrossRef]
- Fahmy, R.G.; Dass, C.R.; Sun, L.Q.; Chesterman, C.N.; Khachigian, L.M. Transcription factor Egr-1 supports FGF-dependent angiogenesis during neovascularization and tumor growth. Nat. Med. 2003, 9, 1026–1032. [Google Scholar] [CrossRef]
- Yang, L.; Xiao, L.; Ma, X.; Tang, M.; Weng, X.; Chen, X.; Sun, L.; Cao, Y. Effect of DNAzymes targeting akt1 on cell proliferation and apoptosis in nasopharyngeal carcinoma. Cancer Biol. Ther. 2009, 8, 366–371. [Google Scholar]
- Bhindi, R.; Fahmy, R.G.; Lowe, H.C.; Chesterman, C.N.; Dass, C.R.; Cairns, M.J.; Saravolac, E.G.; Sun, L.Q.; Khachigian, L.M. Brothers in arms: DNA enzymes, short interfering RNA, and the emerging wave of small-molecule nucleic acid-based gene-silencing strategie. Am. J. Pathol. 2007, 171, 1079–1088. [Google Scholar] [CrossRef]
- Fielding, C.A.; Sandvej, K.; Mehl, A.; Brennan, P.; Jones, M.; Rowe, M. Epstein-Barr virus lmp-1 natural sequence variants differ in their potential to activate cellular signaling pathways. J. Virol. 2001, 75, 9129–9141. [Google Scholar] [CrossRef]
- Cairns, M.J.; Hopkins, T.M.; Witherington, C.; Wang, L.; Sun, L.Q. Target site selection for an RNA-cleaving catalytic DNA. Nat. Biotechnol. 1999, 17, 480–486. [Google Scholar] [CrossRef]
- Cairns, M.J.; King, A.; Sun, L.Q. Optimisation of the 10-23 DNAzyme-substrate pairing interactions enhanced RNA cleavage activity at purine-cytosine target sites. Nucl. Acids Res. 2003, 31, 2883–2889. [Google Scholar] [CrossRef]
- Nunamaker, E.A.; Zhang, H.Y.; Shirasawa, Y.; Benoit, J.N.; Dean, D.A. Electroporation-mediated delivery of catalytic oligodeoxynucleotides for manipulation of vascular gene expression. Am. J. Physiol. Heart Circ. Physiol. 2003, 285, H2240–H2247. [Google Scholar]
- Lu, Z.X.; Ye, M.; Yan, G.R.; Li, Q.; Tang, M.; Lee, L.M.; Sun, L.Q.; Cao, Y. Effect of EBV LMP11 targeted DNAzymes on cell proliferation and apoptosis. Cancer Gene Ther. 2005, 12, 647–654. [Google Scholar] [CrossRef]
- Dass, C.R.; Saravolac, E.G.; Li, Y.; Sun, L.Q. Cellular uptake, distribution, and stability of 10-23 deoxyribozymes. Antisense Nucleic Acid Drug Dev. 2002, 12, 289–299. [Google Scholar] [CrossRef]
- Benimetskaya, L.; Takle, G.B.; Vilenchik, M.; Lebedeva, I.; Miller, P.; Stein, C.A. Cationic porphyrins: Novel delivery vehicles for antisense oligodeoxynucleotides. Nucl. Acids Res. 1998, 26, 5310–5317. [Google Scholar] [CrossRef]
- Lu, Z.X.; Ma, X.Q.; Yang, L.F.; Wang, Z.L.; Zeng, L.; Li, Z.J.; Li, X.N.; Tang, M.; Yi, W.; Gong, J.P.; Sun, L.Q.; Cao, Y. DNAzymes targeted to EBV-encoded latent membrane protein-1 induce apoptosis and enhance radiosensitivity in nasopharyngeal carcinoma. Cancer Lett. 2008, 265, 226–238. [Google Scholar] [CrossRef]
- Yoshizaki, T.; Sato, H.; Furukawa, M.; Pagano, J.S. The expression of matrix metalloproteinase 9 is enhanced by Epstein-Barr virus latent membrane protein 1. Proc. Natl. Acad Sci. USA 1998, 95, 3621–3626. [Google Scholar]
- Rubin, C.I.; Atweh, G.F. The role of stathmin in the regulation of the cell cycle. J. Cell Biochem. 2004, 93, 242–250. [Google Scholar] [CrossRef]
- Mistry, S.J.; Atweh, G.F. Role of stathmin in the regulation of the mitotic spindle: Potential applications in cancer therapy. Mt. Sinai J. Med. 2002, 69, 299–304. [Google Scholar]
- Lin, X.; Liu, S.; Luo, X.; Ma, X.; Guo, L.; Li, L.; Li, Z.; Tao, Y.; Cao, Y. EBV-encoded LMP1 regulates op18/stathmin signaling pathway by cdc2 mediation in nasopharyngeal carcinoma cells. Int. J. Cancer 2009, 124, 1020–1027. [Google Scholar] [CrossRef]
- Korotayev, K.; Chaussepied, M.; Ginsberg, D. Erk activation is regulated by E2F1 and is essential for E2F1-induced s phase entry. Cell. Signal. 2008, 20, 1221–1226. [Google Scholar] [CrossRef]
- Ding, L.; Li, L.; Yang, J.; Zhou, S.; Li, W.; Tang, M.; Shi, Y.; Yi, W.; Cao, Y. Latent membrane protein 1 encoded by Epstein-Barr virus induces telomerase activity via p16ink4a/rb/e2f1 and jnk signaling pathways. J. Med. Virol. 2007, 79, 1153–1163. [Google Scholar] [CrossRef]
- Fan, M.; Ahmed, K.M.; Coleman, M.C.; Spitz, D.R.; Li, J.J. Nuclear factor-kappaB and manganese superoxide dismutase mediate adaptive radioresistance in low-dose irradiated mouse skin epithelial cells. Cancer Res. 2007, 67, 3220–3228. [Google Scholar] [CrossRef]
- Yin, L.; Liao, W.; Deng, X.; Tang, M.; Gu, H.; Li, X.; Yi, W.; Cao, Y. LMP1 activates NF-kappa B via degradation of I kappa B alpha in nasopharyngeal carcinoma cells. Chin. Med. J. (Engl.) 2001, 114, 718–722. [Google Scholar]
- Sommer, S.S.; Jiang, Z.; Feng, J.; Buzin, C.H.; Zheng, J.; Longmate, J.; Jung, M.; Moulds, J.; Dritschilo, A. ATM missense mutations are frequent in patients with breast cancer. Cancer Genet. Cytogenet. 2003, 145, 115–120. [Google Scholar] [CrossRef]
- Pandita, T.K. A multifaceted role for ATM in genome maintenance. Expert Rev. Mol. Med. 2003, 5, 1–21. [Google Scholar]
- Liu, Y.P.; Tan, Y.N.; Wang, Z.L.; Zeng, L.; Lu, Z.X.; Li, L.L.; Luo, W.; Tang, M.; Cao, Y. Phosphorylation and nuclear translocation of STAT3 regulated by the Epstein-Barr virus latent membrane protein 1 in nasopharyngeal carcinoma. Int. J. Mol. Med. 2008, 21, 153–162. [Google Scholar]
- Lo, K.W.; To, K.F.; Huang, D.P. Focus on nasopharyngeal carcinoma. Cancer Cell 2004, 5, 423–428. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds are available from the authors.
© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yang, L.; Lu, Z.; Ma, X.; Cao, Y.; Sun, L.-Q. A Therapeutic Approach to Nasopharyngeal Carcinomas by DNAzymes Targeting EBV LMP-1 Gene. Molecules 2010, 15, 6127-6139. https://doi.org/10.3390/molecules15096127
Yang L, Lu Z, Ma X, Cao Y, Sun L-Q. A Therapeutic Approach to Nasopharyngeal Carcinomas by DNAzymes Targeting EBV LMP-1 Gene. Molecules. 2010; 15(9):6127-6139. https://doi.org/10.3390/molecules15096127
Chicago/Turabian StyleYang, Lifang, Zhongxin Lu, Xiaoqian Ma, Ya Cao, and Lun-Quan Sun. 2010. "A Therapeutic Approach to Nasopharyngeal Carcinomas by DNAzymes Targeting EBV LMP-1 Gene" Molecules 15, no. 9: 6127-6139. https://doi.org/10.3390/molecules15096127



