Transcriptional and Functional Analysis Shows Sodium Houttuyfonate-Mediated Inhibition of Autolysis in Staphylococcus aureus
Abstract
:1. Intrduction
2. Results
2.1. Antimicrobial Activities of SH and Growth Curve of S. aureus under SH Stress
| Strains | Source | Date of isolation (mo/yr) | Antibiotic resistances | VAN MIC (µg/mL) | CIP MIC (µg/mL) | OX MIC (µg/mL) | SH MIC (µg/mL) | SH MBIC (µg/mL) | SH MBBC (µg/mL) |
|---|---|---|---|---|---|---|---|---|---|
| SA003 | Blood | 2/2003 | P | 1 | 0.12 | 0.12 | 4 | >1024 | >1024 |
| SA006 | Skin | 2/2003 | M, P, CI, CM, E | 1 | 64 | 128 | 16 | >1024 | >1024 |
| SA009 | Skin abscess | 3/2003 | M, CI, CM, E | 0.5 | 128 | 256 | 16 | >1024 | >1024 |
| SA017 | Skin abscess | 5/2003 | P, T | 2 | 0.5 | 0.5 | 4 | >1024 | >1024 |
| SA018 | Broncheal swab | 6/2003 | M, P, CI, CM, E, G | 2 | 128 | 512 | 32 | >1024 | >1024 |
| SA025 | Sputum | 10/2003 | — | 0.25 | 0.25 | 0.12 | 4 | >1024 | >1024 |
| SA039 | Broncheal swab | 2/2004 | M, P, CI, CM, E, G, RI, T, TMP/SXT | 1 | 512 | 512 | 128 | >1024 | >1024 |
| SA059 | Broncheal swab | 6/2004 | M, P, CI, T, TMX/SXT | 2 | 128 | 512 | 32 | >1024 | >1024 |
| SA079 | Sputum | 9/2004 | M, P, CI, CL, CM, E, G, TMP/SXT | 4 | 128 | 256 | 64 | >1024 | >1024 |
| SA092 | Abscess | 10/2004 | M, P, OX, E | 1 | 16 | 128 | 32 | >1024 | >1024 |
| SA106 | Wound isolate | 1/2005 | M, P, CI, CM, E, G, T, TMP/SXT | 0.5 | 256 | 512 | 32 | >1024 | >1024 |
| SA118 | Blood | 1/2005 | M, P, E, G | 1 | 64 | 256 | 8 | >1024 | >1024 |
| SA121 | Wound isolate | 3/2005 | M, P, CM, E, G | 1 | 128 | 256 | 16 | >1024 | >1024 |
| SA142 | Abscess | 5/2005 | P, T, E | 1 | 1 | 0.5 | 8 | >1024 | >1024 |
| SA146 | Skin abscess | 5/2005 | M, P, CL, CM, E, T | 2 | 64 | 128 | 16 | >1024 | >1024 |
| SA165 | Urine | 6/2006 | P, CI, CM, E, G, RI, T | 2 | 256 | 512 | 32 | >1024 | >1024 |
| SA173 | Broncheal swab | 8/2005 | M, P, T | 0.25 | 32 | 128 | 8 | >1024 | >1024 |
| SA179 | Blood | 9/2005 | M, CI, CM, E | 1 | 128 | 512 | 32 | >1024 | >1024 |
| SA192 | Urine | 10/2005 | M, P, CI, CM, E, G | 1 | 64 | 512 | 32 | >1024 | >1024 |
| SA203 | Wound isolate | 12/2005 | M, P, CL, CM, T | 2 | 64 | 512 | 16 | >1024 | >1024 |
| ATCC25923 | CMCC | — | 1 | 0.25 | 0.25 | 16 | >1024 | >1024 |

2.2. Overview of SH-Triggered Transcriptional Profiles of S. aureus ATCC 25923 Cells
2.3. Expression Levels of Autolysis-Associated Genes Treated by SH
| N315 SA no. | Gene | Description | Microarray a,c | RT-PCR b,c,d | |
|---|---|---|---|---|---|
| Fold change ± SD | q value (%) | Fold change ± SD | |||
| SA0905 | atl | Bifunctional precursor autolysin (Atl) | −2.2 ± 0.5 ** | 0.11 | −2.4 ± 0.8 § |
| SA0423 | sle1 | N-Acetylmuramyl-L-alanine amidase | −12.4 ± 3.0 ** | 0.08 | −31.2 ± 9.6 §§ |
| SA2329 | cidA | Hypothetical protein, similar to transcription regulator | −2.0 ± 0.6 ** | 0.38 | −3.7 ± 0.5 § |
| SA0265 | lytM | Peptidoglycan hydrolase | 1.2 ± 0.6 | 29.61 | 1.0 ± 0.6 |
| SA1090 | lytN | LytN protein | −1.4 ± 0.5 | 9.33 | −2.2 ± 0.9 § |
| SA0252 | lrgA | Murein hydrolase regulator LrgA | 24.2 ± 5.2 ** | 0.00 | 80.4 ± 18.9 §§ |
| SA0253 | lrgB | Antiholin-like protein LrgB | 21.5 ± 6.4 ** | 0.00 | 76.5 ±14.7 §§ |
| SA0573 | sarA | Staphylococcal accessory regulator A | 1.8 ± 0.7 ** | 2.69 | 2.6 ± 0.9 § |
| SA2328 | cidB | Conserved hypothetical protein | 1.5 ± 0.4 * | 4.53 | 2.5 ± 1.0 §§ |
| SA2327 | cidC | Pyruvate oxidase | 2.4 ± 0.5 ** | 0.13 | 2.3 ±0.7 § |
| SA1844 | agrA | Accessory gene regulator A | −2.2 ± 0.7 ** | 0.19 | −3.5 ± 0.6 §§ |
| SAS065 | RNAIII | Delta hemolysin | −8.3 ± 1.4 ** | 0.00 | −21.9 ± 5.3 §§ |
| SA0251 | lytR | Two-component response regulator | −3.9 ± 0.9 ** | 0.00 | −9.1 ± 1.4 §§ |
| SA0250 | lytS | Two-component sensor histidine kinase | −1.9 ± 0.5 ** | 0.25 | −2.4 ± 1.1 § |
| SA0641 | mgrA | Transcriptional regulator MgrA | −2.3 ± 0.8 ** | 0.11 | −3.2 ± 0.9 §§ |
| SA1246 | arlS | Sensor histidine kinase ArlS | −1.6 ± 0.4 ** | 0.83 | −2.2 ± 0.7 § |
| SA1248 | arlR | Truncated (putative response regulator ArlR | −1.1 ± 0.5 | 29.61 | −1.2 ± 0.5 |
| SA0650 | norA | Quinolone resistance protein | 1.2 ± 0.38 | 27.61 | ND |
| SA0904 | Hypothetical protein, probable ATL autolysin transcription regulator | −1.5 ± 0.11 * | 3.70 | ND | |
| SA1091 | eprH | Endopeptidase resistance gene | −1.3 ± 0.10 | 13.98 | ND |
| Primer | N315 ORF a | Sequence |
|---|---|---|
| 16S rRNA for | SArRNA01 | CGTGCTACAATGGACAATACAAA |
| 16S rRNA rev | SArRNA01 | ATCTACGATTACTAGCGATTCCA |
| atl for | N315-SA0905 | TACCGTAACGGCGTAGGTCGT |
| atl rev | N315-SA0905 | CATAGTCGTGTGTGTGTACGA |
| Sle1for | N315-SA0423 | GTAGCCGTCCATCAACGAACT |
| Sle1 rev | N315-SA0423 | CTATTGCTCGCAGCGTTACT |
| cidA for | N315-SA2329 | CTTAGCCGGCAGTATTGTTG |
| cidA rev | N315-SA2329 | TGAAGATAATGCAACGATAC |
| lytM for | N315-SA0265 | ATGCCAATGGAAGCGGCCA |
| lytM rev | N315-SA0265 | TTCGCATGACCACTAGCTGT |
| lytN for | N315-SA1090 | GGAGACACACTTAGTGCT |
| lytN rev | N315-SA1090 | CTAATGGTGTCATTGGCACT |
| lrgA for | N315-SA0252 | CTGGTGCTGTTAAGTTAGGCG |
| lrgA rev | N315-SA0252 | GTGACATAGCCAGTACAAAT |
| lrgB for | N315-SA0253 | CGGTACAGTTGTAGCGTTATTA |
| lrgB rev | N315-SA0253 | AGTGCTAATCCTCGGGCAATA |
| sarA for | N315-SA0573 | CGCTGTATTGACATACATCAGCG |
| sarA rev | N315-SA0573 | CTCGACTCAATAATGATTCGA |
| cidB for | N315-SA2328 | GACGTCATTGTAACGTTATTGC |
| cidB rev | N315-SA2328 | TGAACTAAATGCACCGGATTC |
| cidC for | N315-SA2327 | GAGACTCAATCGACGCAGTTGT |
| cidC rev | N315-SA2327 | TCCAAGTGCTGTACTATTCG |
| agrA for | N315-SA1844 | TGAAATTCGTAAGCATGACCC |
| agrA rev | N315-SA1844 | CATCGCTGCAACTTTGTAGAC |
| RNAIII for | N315-SAS065 | ATGAGTTGTTTAATTTTAAGAAT |
| RNAIII rev | N315-SAS065 | CACTGTGTCGATAATCCA |
| lytS for | N315-SA0250 | GCATATGCGGTTGACCTCATAT |
| lytS rev | N315-SA0250 | GCTTGTAATTGCTACGGCAGA |
| mgrA for | N315-SA0641 | CAAGTTAATCGCTACTACTC |
| mgrA rev | N315-SA0641 | CTTCACGTTGATCGACTTCG |
| arlS for | N315-SA1246 | GTATATCATTGCGCTGGCA |
| arlS rev | N315-SA1246 | GTGTTCGTAATTCATGTGACGC |
2.4. SH Decreases Triton X-100-Induced Autolysis
2.5. SH Alters the Cellular Peptidoglycan Hydrolase Profile


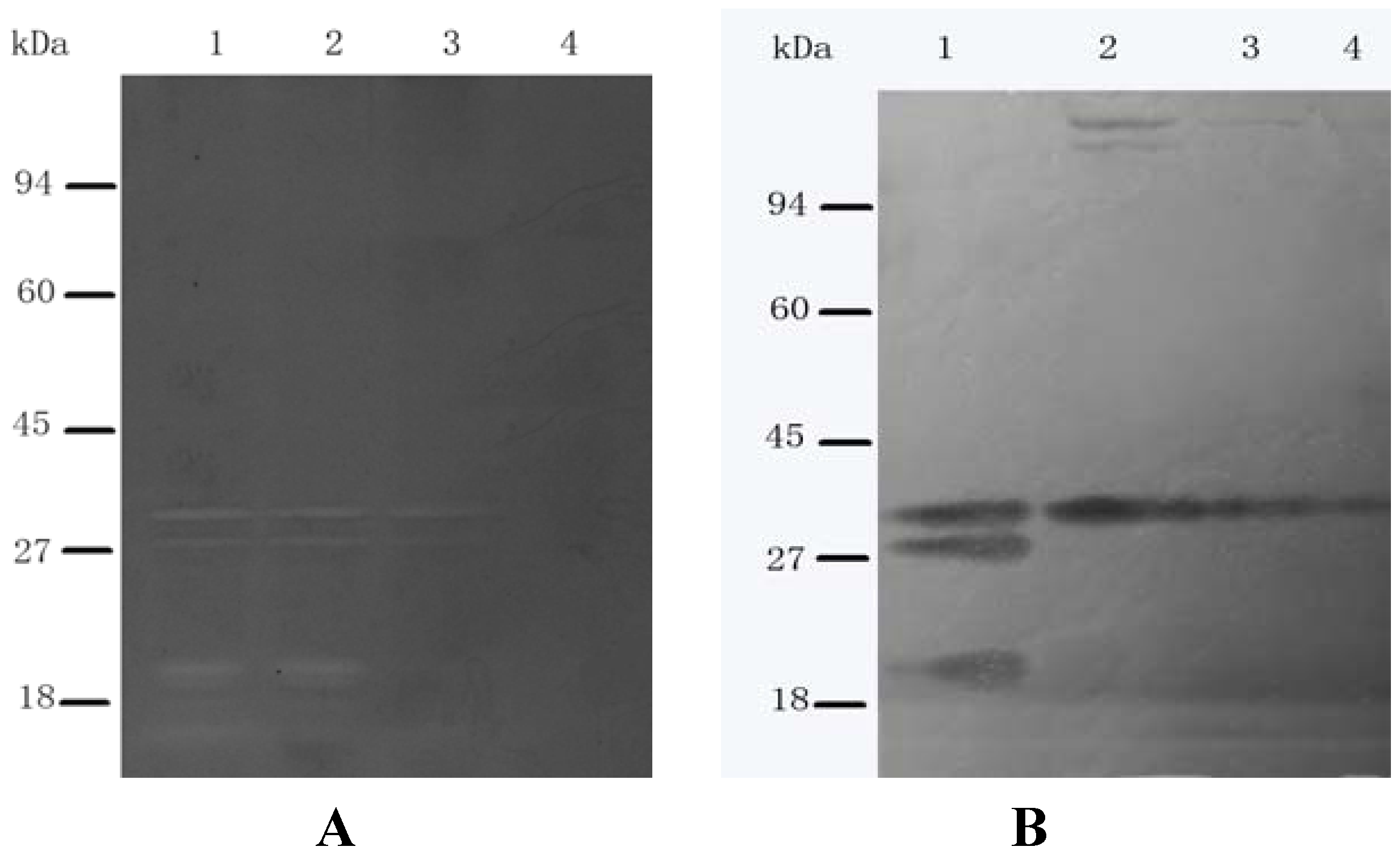
2.6. Role of SH on DNA Release by S. aureus

3. Discussion
4. Experimental
4.1. Bacterial Strains and Materials
4.2. Antibiotic Susceptibility Testing
4.3. Growth Curves
4.4. Treatment with SH for Microarrays
4.5. RNA Isolation and cDNA Labeling
4.6. Quantitative Real-Time RT-PCR
4.7. Triton X-100-Induced Autolysis
4.8. Quantitative Bacteriolytic Hydrolase Assays
4.9. Autolytic Enzyme Extracts
4.10. Preparation of Crude Cell Walls
4.11. Bacteriolytic Enzyme Profiles after SDS-Polyacrylamide Gel Electrophoresis
4.12. Establishment of Microbial Biofilms
4.13. Biofilm Antimicrobial Susceptibility Testing
4.14. Measurement of eDNA in Microtiter Plate Cultures Treated with SH
5. Conclusion
Supplementary Materials
Supplementary Materials
Supplementary File 1Acknowledgements
Conflict of interest
Reference
- Paulsen, I.T.; Firth, M.; Skurray, R.A. Resistance to Antimicrobial Agents other than b-Lactams. In The Staphylococci in Human Disease; Crossley, K.B., Archer, G.L., Eds.; Churchill Livingstone: New York, NY, USA, 1997; pp. 175–212. [Google Scholar]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef]
- Parsek, M.R.; Fuqua, C. Biofilms 2003: Emerging themes and challenges in studies of surface-associated microbial life. J. Bacteriol. 2004, 186, 4427–4440. [Google Scholar] [CrossRef]
- Wang, D.; Yu, Q.; Eikstadt, P.; Hammond, D.; Feng, Y.; Chen, N. Studies on adjuvanticity of sodium houttuyfonate and its mechanism. Int. Immunopharmacol. 2002, 2, 1411–1418. [Google Scholar] [CrossRef]
- The Pharmacopoeia Committee of China, The Chinese Pharmacopoeia, vol. I; The Pharmacopoeia Committee of China, The Chinese Pharmacopoeia, vol. I: Beijing, China, 2000.
- Li, S.; Yu, Q.H.; Chu, Z.Y. Studies on the anti-inflammatory effect and mechanism of houttuyninum. Chin. Pharmacol. Bull. 2007, 14, 442–444. [Google Scholar]
- Ye, X.L.; Li, X.G.; Yuan, L.J.; Ge, L.H.; Zhang, B.S.; Zhou, S.B. Interaction of houttuyfonate homologues with the cell membrane of gram-positive and gram-negative bacteria. Colloids Surf. A 2007, 301, 412–418. [Google Scholar] [CrossRef]
- Rice, K.C.; Mann, E.E.; Endres, J.L.; Weiss, E.C.; Cassat, J.E. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2007, 104, 8113–8118. [Google Scholar]
- Reinicke, B.; Blumel, P.; Giesbrecht, P. Reduced degradability by lysozyme of staphylococcal cell walls after chloramphenicol treatment. Arch. Microbiol. 1983, 135, 120–124. [Google Scholar] [CrossRef]
- Yamada, S.; Sugai, M.; Komatsuzawa, H.; Matsumoto, A. Suppressed localization of a major autolysin on Staphylococcus aureus treated with tetracycline. J. Electron. Microsc. 2001, 50, 359–364. [Google Scholar] [CrossRef]
- Sieradzki, K.; Tomasz, A. Inhibition of the autolytic system by vancomycin causes mimicry of vancomycin-intermediate Staphylococcus aureus-type resistance, cell concentration dependence of the MIC, and antibiotic tolerance in vancomycin-susceptible S. aureus. Antimicrob. Agents Chemother. 2006, 50, 527–533. [Google Scholar] [CrossRef]
- Yoshida, R.; Kuwahara-Arai, K.; Baba, T.; Cui, L.; Richardson, J.F.; Hiramatsu, K. Physiological and molecular analysis of a mecA-negative Staphylococcus aureus clinical strain that expresses heterogeneous methicillin resistance. J. Antimicrob. Chemother. 2003, 51, 247–255. [Google Scholar] [CrossRef]
- Ledala, N.; Wilkinson, B.J.; Jayaswal, R.K. Effects of oxacillin and tetracycline on autolysis, autolysin processing and atl transcription in Staphylococcus aureus. Int. J. Antimicrob. Agents 2006, 27, 518–524. [Google Scholar] [CrossRef]
- Michel, A.; Agerer, F.; Hauck, C.R.; Herrmann, M.; Ullrich, J.; Hacker, J.; Ohlsen, K. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repar. J. Bacteriol. 2006, 188, 5783–5796. [Google Scholar] [CrossRef]
- Liang, X.; Zheng, L.; Landwehr, C.; Lunsford, D.; Holmes, D.; Ji, Y. Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J. Bacteriol. 2005, 187, 5486–5492. [Google Scholar] [CrossRef]
- Rice, K.C.; Patton, T.; Yang, S.J.; Dumoulin, A.; Bischoff, M.; Bayles, K.W. Transcription of the Staphylococcus aureus cid and lrg murein hydrolase regulators is affected by sigma factor B. J. Bacteriol. 2004, 186, 3029–3037. [Google Scholar] [CrossRef]
- Dubrac, S.; Boneca, I.G.; Poupel, O.; Msadek, T. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 2007, 189, 8257–8269. [Google Scholar] [CrossRef]
- Rice, K.; Peralta, R.; Bast, D.; de Azavedo, J.; McGavin, M.J. Description of Staphylococcus serine protease (ssp) operon in Staphylococcus aureus and nonpolar inactivation of sspA-encoded serine protease. Infect. Immun. 2001, 69, 159–169. [Google Scholar] [CrossRef]
- Ingavale, S.S.; van Wamel, W.; Cheung, A.L. Characterization of RAT, an autolysis regulator in Staphylococcus aureus. Mol. Microbiol. 2003, 48, 1451–1466. [Google Scholar] [CrossRef]
- Manna, A.C.; Ingavale, S.S.; Maloney, M.; van Wamel, W.; Cheung, A.L. Identification of sarV (SA2062), a new transcriptional regulator, is repressed by SarA and MgrA (SA0641) and involved in the regulation of autolysis in Staphylococcus aureus. J. Bacteriol. 2004, 186, 5267–5280. [Google Scholar] [CrossRef]
- Rice, K.C.; Patton, T.; Yang, S.J.; Dumoulin, A.; Bischoff, M.; Bayles, K.W. Transcription of the Staphylococcus aureuscid and lrg murein hydrolase regulators is affected by sigma factor B. J. Bacteriol. 2004, 186, 3029–3037. [Google Scholar] [CrossRef]
- Wootton, M.; Bennett, P.M.; MacGowan, A.P.; Walsh, T.R. Reduced expression of the atl autolysin gene and susceptibility to autolysis in clinical heterogeneous glycopeptide-intermediate Staphylococcus aureus (hGISA) and GISA strains. J. Antimicrob. Chemother. 2005, 56, 944–947. [Google Scholar] [CrossRef]
- Saïd-Salim, B.; Dunman, P.M.; McAleese, F.M.; Macapagal, D.; Murphy, E.; McNamara, P.J.; Arvidson, S.; Foster, T.J.; Projan, S.J.; Kreiswirth, B.N. Global Regulation of Staphylococcus aureus Genes by Rot. J. Bacteriol. 2003, 185, 610–619. [Google Scholar] [CrossRef]
- Brunskill, E.W.; Bayles, K.W. Identification of LytSR-regulated genes from Staphylococcus aureus. J. Bacteriol. 1996, 178, 5810–5812. [Google Scholar]
- Gründling, A.; Schneewind, O. Genes required for glycolipid synthesis and lipoteichoic acid anchoring in Staphylococcus aureus. J. Bacteriol. 2007, 189, 2521–2530. [Google Scholar] [CrossRef]
- Brunskill, E.W.; Bayles, K.W. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 1996, 178, 611–618. [Google Scholar]
- Whitchurch, C.B.; Tolker-Nielsen, T.; Ragas, P.C.; Mattick, J.S. Extracellular DNA required for bacterial biofilm formation. Science 2002, 295, 1487. [Google Scholar] [CrossRef]
- Rice, K.C.; Mann, E.E.; Endres, J.L.; Weiss, E.C.; Cassat, J.E. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2007, 104, 8113–8118. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, 8th ed; Approved Standard M7-A8. CLSI: Wayne, PA, USA, 2009.
- Yu, L.; Xiang, H.; Fan, J.; Wang, D.; Yang, F.; Guo, N.; Jin, Q.; Deng, X. Global transcriptional response of Staphylococcus aureus to rhein, a natural plant product. J. Biotechnol. 2008, 135, 304–308. [Google Scholar] [CrossRef]
- McAleese, F.; Wu, S.W.; Sieradzki, K.; Dunman, P.; Murphy, E.; Projan, S.; Tomasz, A. Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate-S. aureus-type resistance to vancomycin. J. Bacteriol. 2006, 188, 1120–1133. [Google Scholar] [CrossRef]
- Tusher, V.G.; Tibshirani, R.; Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 2001, 98, 5116–5121. [Google Scholar] [CrossRef]
- Reddy, S.P.; Britto, R.; Vinnakota, K.; Aparna, H.; Sreepathi, H.K.; Thota, B.; Kumari, A.; Shilpa, B.M.; Vrinda, M.; Umesh, S.; et al. Novel glioblastoma markers with diagnostic and prognostic value identified through transcriptome analysis. Clin. Cancer Res. 2008, 14, 2978–2987. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ray, S.; Chatterjee, E.; Chatterjee, A.; Paul, K.; Chowdhury, R. A fadD mutant of Vibrio cholerae is impaired in the production of virulence factors and membrane localization of the virulence regulatory protein TcpP. Infect. Immun. 2011, 79, 258–266. [Google Scholar] [CrossRef]
- Scaltriti, M.; Belloni, L.; Caporali, A.; Davalli, P.; Remondini, D.; Rizzi, F.; Astancolle, S.; Corti, A.; Bettuzzi, S. Molecular classification of green tea catechin-sensitive and green tea catechin-resistant prostate cancer in the TRAMP mice model by quantitative real-time PCR gene profiling. Carcinogenesis 2006, 27, 1047–1053. [Google Scholar]
- Mani, N.; Tobin, P.; Jayaswal, R.K. Isolation and characterization of autolysis-defective mutants of Staphylococcus aureus created by Tn917-lacZ mutagenesis. J. Bacteriol. 1993, 175, 1493–1499. [Google Scholar]
- Groicher, K.H.; Firek, B.A.; Fujimoto, D.F.; Bayles, K.W. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 2000, 182, 1794–1801. [Google Scholar] [CrossRef]
- Mani, N.; Tobin, P.; Jayaswal, R.K. Isolation and characterization of autolysis-defective mutants of Staphylococcus aureus created by Tn917-lacZ mutagenesis. J. Bacteriol. 1993, 175, 1493–1499. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 277, 680–685. [Google Scholar] [CrossRef]
- Reinicke, B.; Blumel, P.; Giesbrecht, P. Reduced degradability by lysozyme of staphylococcal cell walls after chloramphenicol treatment. Arch. Microbiol. 1983, 135, 120–124. [Google Scholar] [CrossRef]
- Freeman, D.J.; Falkiner, F.R.; Keane, C.T. New method for detecting slime production by coagulase negative staphylococci. J. Clin. Pathol. 1989, 42, 872–874. [Google Scholar] [CrossRef]
- Adams, D.; Quayum, M.; Worthington, T.; Lambert, P.; Elliott, T. Evaluation of a 2% chlorhexidine gluconate in 70% isopropyl alcohol skin disinfectant. J. Hosp. Infect. 2005, 61, 278–290. [Google Scholar]
- Qin, Z.; Ou, Y.; Yang, L.; Zhu, Y.; Tolker-Nielsen, T.; Molin, S.; Qu, D. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 2007, 153, 2083–2092. [Google Scholar] [CrossRef]
- Sample Availability: Sample of the compound Sodium Houttuyfonate is available from the authors.
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Liu, G.; Xiang, H.; Tang, X.; Zhang, K.; Wu, X.; Wang, X.; Guo, N.; Feng, H.; Wang, G.; Liu, L.; et al. Transcriptional and Functional Analysis Shows Sodium Houttuyfonate-Mediated Inhibition of Autolysis in Staphylococcus aureus. Molecules 2011, 16, 8848-8865. https://doi.org/10.3390/molecules16108848
Liu G, Xiang H, Tang X, Zhang K, Wu X, Wang X, Guo N, Feng H, Wang G, Liu L, et al. Transcriptional and Functional Analysis Shows Sodium Houttuyfonate-Mediated Inhibition of Autolysis in Staphylococcus aureus. Molecules. 2011; 16(10):8848-8865. https://doi.org/10.3390/molecules16108848
Chicago/Turabian StyleLiu, Guoxing, Hua Xiang, Xudong Tang, Kaiyu Zhang, Xiuping Wu, Xuelin Wang, Na Guo, Haihua Feng, Guangming Wang, Lihui Liu, and et al. 2011. "Transcriptional and Functional Analysis Shows Sodium Houttuyfonate-Mediated Inhibition of Autolysis in Staphylococcus aureus" Molecules 16, no. 10: 8848-8865. https://doi.org/10.3390/molecules16108848
APA StyleLiu, G., Xiang, H., Tang, X., Zhang, K., Wu, X., Wang, X., Guo, N., Feng, H., Wang, G., Liu, L., Shi, Q., Shen, F., Xing, M., Yuan, P., Liu, M., & Yu, L. (2011). Transcriptional and Functional Analysis Shows Sodium Houttuyfonate-Mediated Inhibition of Autolysis in Staphylococcus aureus. Molecules, 16(10), 8848-8865. https://doi.org/10.3390/molecules16108848




