Oligomeric Nucleic Acids as Antivirals
Abstract
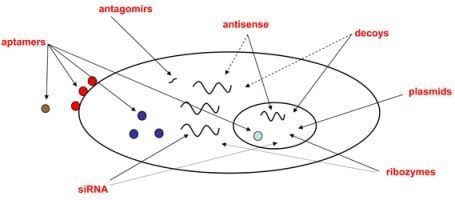
:1. Introduction
3. Advantages of Oligonucleotide-Based Drugs
4. Limitations of Oligonucleotide-Based Drugs

5. Conclusions and Perspectives
| Drug | Class | Virus | Target transcript/s | Developer/Reference | Status |
|---|---|---|---|---|---|
| Fomivirsen (VitraveneTM) | asON | CMV | IE2 | Isis Pharmaceuticals | Approved |
| VRX496 | asON | HIV | env | [32] | Phase II |
| Morpholino asON | asON | ZEBOV and MARV | VP24 and VP35 | [35,36] | Entering Phase I |
| OZ1 | ribozyme | HIV | tat and vpr | [50] | Phase II |
| SPC3649 | anti-miRNA | HCV | miR-122 | Santaris Pharma | Phase I |
| ALN-RSV01 | siRNA | RSV | N-protein transcript | Alnylam | Phase II |
| siRNA-SNALP | siRNA | ZEBOV | L-polymerase, VP24 and VP35 | Tekmira Pharmaceuticals | Pre-clinical |
Acknowledgements
References
- Turner, B.G.; Summers, M.F. Structural biology of HIV. J. Mol. Biol. 1999, 285, 1–32. [Google Scholar] [CrossRef]
- Vivet-Boudou, V.; Didierjean, J.; Isel, C.; Marquet, R. Nucleoside and nucleotide inhibitors of HIV-1 replication. Cell Mol. Life Sci. 2006, 63, 163–186. [Google Scholar] [CrossRef]
- Pomerantz, R.J.; Horn, D.L. Twenty years of therapy for HIV-1 infection. Nat. Med. 2003, 9, 867–873. [Google Scholar] [CrossRef]
- Park, N.H.; Pavan-Langston, D.; McLean, S.L. Acylovir in oral and ganglionic herpes simplex virus infections. J. Infect. Dis. 1979, 140, 802–806. [Google Scholar] [CrossRef]
- Reichard, O.; Andersson, J.; Schvarcz, R.; Weiland, O. Ribavirin treatment for chronic hepatitis C. Lancet 1991, 337, 1058–1061. [Google Scholar] [CrossRef]
- Menendez-Arias, L. Targeting HIV: antiretroviral therapy and development of drug resistance. Trends Pharmacol. Sci. 2002, 23, 381–388. [Google Scholar] [CrossRef]
- Sarafianos, S.G.; Das, K.; Hughes, S.H.; Arnold, E. Taking aim at a moving target: designing drugs to inhibit drug-resistant HIV-1 reverse transcriptases. Curr. Opin. Struct. Biol. 2004, 14, 716–730. [Google Scholar] [CrossRef]
- Turner, D.; Wainberg, M.A. HIV transmission and primary drug resistance. AIDS Rev. 2006, 8, 17–23. [Google Scholar]
- Carr, A. Toxicity of antiretroviral therapy and implications for drug development. Nat. Rev. Drug Discov. 2003, 2, 624–634. [Google Scholar] [CrossRef]
- Pinti, M.; Salomoni, P.; Cossarizza, A. Anti-HIV drugs and the mitochondria. Biochim. Biophys. Acta 2006, 1757, 700–707. [Google Scholar]
- Nimjee, S.M.; Rusconi, C.P.; Sullenger, B.A. Aptamers: an emerging class of therapeutics. Annu. Rev. Med. 2005, 56, 555–583. [Google Scholar]
- Blank, M.; Blind, M. Aptamers as tools for target validation. Curr. Opin. Chem. Biol. 2005, 9, 336–342. [Google Scholar] [CrossRef]
- Gopinath, S.C. Antiviral aptamers. Arch. Virol. 2007, 152, 2137–2157. [Google Scholar] [CrossRef]
- Pan, Q.W.; Henry, S.D.; Scholte, B.J.; Tilanus, H.W.; Janssen, H.L.; van der Laan, L.J. New therapeutic opportunities for hepatitis C based on small RNA. World J. Gastroenterol. 2007, 13, 4431–4436. [Google Scholar]
- Haasnoot, J.; Berkhout, B. Nucleic acids-based therapeutics in the battle against pathogenic viruses. Handb. Exp. Pharmacol. 2009, 189, 243–263. [Google Scholar] [CrossRef]
- Zamecnik, P.C.; Stephenson, M.L. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc. Natl. Acad. Sci. USA 1978, 75, 280–284. [Google Scholar] [CrossRef]
- Stephenson, M.L.; Zamecnik, P.C. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc. Natl. Acad. Sci. USA 1978, 75, 285–288. [Google Scholar] [CrossRef]
- Kurreck, J. Antisense technologies. Improvement through novel chemical modifications. Eur. J. Biochem. 2003, 270, 1628–1644. [Google Scholar] [CrossRef]
- Corey, D.R. RNA learns from antisense. Nat. Chem. Biol. 2007, 3, 8–11. [Google Scholar] [CrossRef]
- Bennett, C.F.; Swayze, E.E. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 259–293. [Google Scholar]
- Sczakiel, G.; Far, R.K. The role of target accessibility for antisense inhibition. Curr. Opin. Mol. Ther. 2002, 4, 149–153. [Google Scholar]
- Pan, W.H.; Clawson, G.A. Identifying accessible sites in RNA: the first step in designing antisense reagents. Curr. Med. Chem. 2006, 13, 3083–3103. [Google Scholar] [CrossRef]
- Sahu, N.K.; Shilakari, G.; Nayak, A.; Kohli, D.V. Antisense technology: a selective tool for gene expression regulation and gene targeting. Curr. Pharm. Biotechnol. 2007, 8, 291–304. [Google Scholar] [CrossRef]
- Patzel, V.; Steidl, U.; Kronenwett, R.; Haas, R.; Sczakiel, G. A theoretical approach to select effective antisense oligodeoxyribonucleotides at high statistical probability. Nucl. Acid. Res. 1999, 27, 4328–4334. [Google Scholar] [CrossRef]
- Far, R.K.; Leppert, J.; Frank, K.; Sczakiel, G. Technical improvements in the computational target search for antisense oligonucleotides. Oligonucleotides 2005, 15, 223–233. [Google Scholar]
- Spurgers, K.B.; Sharkey, C.M.; Warfield, K.L.; Bavari, S. Oligonucleotide antiviral therapeutics: antisense and RNA interference for highly pathogenic RNA viruses. Antivir. Res. 2008, 78, 26–36. [Google Scholar]
- Hnik, P.; Boyer, D.S.; Grillone, L.R.; Clement, J.G.; Henry, S.P.; Green, E.A. Antisense oligonucleotide therapy in diabetic retinopathy. J. Diabetes Sci. Technol. 2009, 3, 924–930. [Google Scholar]
- Seguin, R.M.; Ferrari, N. Emerging oligonucleotide therapies for asthma and chronic obstructive pulmonary disease. Expert Opin. Invest. Drugs 2009, 18, 1505–1517. [Google Scholar] [CrossRef]
- Aartsma-Rus, A. Antisense-mediated modulation of splicing: Therapeutic implications for duchenne muscular dystrophy. RNA Biol. 2010, 7, 453–461. [Google Scholar] [CrossRef]
- Geary, R.S.; Henry, S.P.; Grillone, L.R. Fomivirsen: clinical pharmacology and potential drug interactions. Clin. Pharmacokinet. 2002, 41, 255–260. [Google Scholar] [CrossRef]
- Schreiber, A.; Harter, G.; Schubert, A.; Bunjes, D.; Mertens, T.; Michel, D. Antiviral treatment of cytomegalovirus infection and resistant strains. Expert Opin. Pharmacother. 2009, 10, 191–209. [Google Scholar] [CrossRef]
- Levine, B.L.; Humeau, L.M.; Boyer, J.; MacGregor, R.R.; Rebello, T.; Lu, X.; Binder, G.K.; Slepushkin, V.; Lemiale, F.; Mascola, J.R.; Bushman, F.D.; Dropulic, B.; June, C.H. Gene transfer in humans using a conditionally replicating lentiviral vector. Proc. Natl. Acad. Sci. USA 2006, 103, 17372–17377. [Google Scholar]
- Matzen, K.; Elzaouk, L.; Matskevich, A.A.; Nitzsche, A.; Heinrich, J.; Moelling, K. RNase H-mediated retrovirus destruction in vivo triggered by oligodeoxynucleotides. Nat. Biotechnol. 2007, 25, 669–674. [Google Scholar] [CrossRef]
- Witherell, G.W. ISIS-14803 (Isis Pharmaceuticals). Curr. Opin. Invest. Drugs 2001, 2, 1523–1529. [Google Scholar]
- Swenson, D.L.; Warfield, K.L.; Warren, T.K.; Lovejoy, C.; Hassinger, J.N.; Ruthel, G.; Blouch, R.E.; Moulton, H.M.; Weller, D.D.; Iversen, P.L.; Bavari, S. Chemical modifications of antisense morpholino oligomers enhance their efficacy against Ebola virus infection. Antimicrob. Agents Chemother. 2009, 53, 2089–2099. [Google Scholar]
- Warren, T.K.; Warfield, K.L.; Wells, J.; Swenson, D.L.; Donner, K.S.; Van Tongeren, S.A.; Garza, N.L.; Dong, L.; Mourich, D.V.; Crumley, S.; Nichols, D.K.; Iversen, P.L.; Bavari, S. Advanced antisense therapies for postexposure protection against lethal filovirus infections. Nat. Med. 2010, 16, 991–994. [Google Scholar] [CrossRef]
- Lilley, D.M. Structure, folding and mechanisms of ribozymes. Curr. Opin. Struct. Biol. 2005, 15, 313–323. [Google Scholar] [CrossRef]
- Walter, N.G. Ribozyme catalysis revisited: is water involved? Mol. Cell 2007, 28, 923–929. [Google Scholar] [CrossRef]
- Serganov, A.; Patel, D.J. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat. Rev. Genet. 2007, 8, 776–790. [Google Scholar] [CrossRef]
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef]
- Chen, X.; Li, N.; Ellington, A.D. Ribozyme catalysis of metabolism in the RNA world. Chem. Biodivers. 2007, 4, 633–655. [Google Scholar] [CrossRef]
- Strobel, S.A.; Cochrane, J.C. RNA catalysis: ribozymes, ribosomes, and riboswitche. Curr. Opin. Chem. Biol. 2007, 11, 636–643. [Google Scholar] [CrossRef]
- Peracchi, A. Prospects for antiviral ribozymes and deoxyribozymes. Rev. Med. Virol. 2004, 14, 47–64. [Google Scholar] [CrossRef]
- Opalinska, J.B.; Gewirtz, A.M. Nucleic-acid therapeutics: basic principles and recent applications. Nat. Rev. Drug Discov. 2002, 1, 503–514. [Google Scholar] [CrossRef]
- Sullenger, B.A.; Gilboa, E. Emerging clinical applications of RNA. Nature 2002, 418, 252–258. [Google Scholar] [CrossRef]
- Sakamoto, N.; Wu, C.H.; Wu, G.Y. Intracellular cleavage of hepatitis C virus RNA and inhibition of viral protein translation by hammerhead ribozymes. J. Clin. Invest. 1996, 98, 2720–2728. [Google Scholar] [CrossRef]
- Ryu, K.J.; Lee, S.W. Identification of the most accessible sites to ribozymes on the hepatitis C virus internal ribosome entry site. J. Biochem. Mol. Biol. 2003, 36, 538–544. [Google Scholar] [CrossRef]
- Trepanier, J.B.; Tanner, J.E.; Alfieri, C. Oligonucleotide-based therapeutic options against hepatitis C virus infection. Antivir. Ther. 2006, 11, 273–287. [Google Scholar]
- Levesque, M.V.; Levesque, D.; Briere, F.P.; Perreault, J.P. Investigating a new generation of ribozymes in order to target HCV. PLoS One 2010, 5, e9627. [Google Scholar]
- Mitsuyasu, R.T.; Merigan, T.C.; Carr, A.; Zack, J.A.; Winters, M.A.; Workman, C.; Bloch, M.; Lalezari, J.; Becker, S.; Thornton, L.; Akil, B.; Khanlou, H.; Finlayson, R.; McFarlane, R.; Smith, D.E.; Garsia, R.; Ma, D.; Law, M.; Murray, J.M.; von Kalle, C.; Ely, J.A.; Patino, S.M.; Knop, A.E.; Wong, P.; Todd, A.V.; Haughton, M.; Fuery, C.; Macpherson, J.L.; Symonds, G.P.; Evans, L.A.; Pond, S.M.; Cooper, D.A. Phase 2 gene therapy trial of an anti-HIV ribozyme in autologous CD34+ cells. Nat. Med. 2009, 15, 285–292. [Google Scholar]
- Burnett, J.C.; Rossi, J.J. Stem cells, ribozymes and HIV. Gene Ther. 2009, 16, 1178–1179. [Google Scholar] [CrossRef]
- Müller-Kuller, T.; Capalbo, G.; Klebba, C.; Engels, J.W.; Klein, S.A. Identification and characterization of a highly efficient anti-HIV pol hammerhead ribozyme. Oligonucleotides 2009, 19, 265–272. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar]
- Cullen, B.R. Five questions about viruses and microRNAs. PLoS Pathog. 2010, 6, e1000787. [Google Scholar] [CrossRef]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef]
- Schwarz, D.S.; Hutvagner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef]
- Okamura, K.; Phillips, M.D.; Tyler, D.M.; Duan, H.; Chou, Y.T.; Lai, E.C. The regulatory activity of microRNA* species has substantial influence on microRNA and 3' UTR evolution. Nat. Struct. Mol. Biol. 2008, 15, 354–363. [Google Scholar] [CrossRef]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar]
- Castanotto, D.; Rossi, J.J. The promises and pitfalls of RNA-interference-based therapeutics. Nature 2009, 457, 426–433. [Google Scholar] [CrossRef]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef]
- Mack, G.S. MicroRNA gets down to business. Nat. Biotechnol. 2007, 25, 631–638. [Google Scholar] [CrossRef]
- Mattes, J.; Yang, M.; Foster, P.S. Regulation of microRNA by antagomirs: a new class of pharmacological antagonists for the specific regulation of gene function? Am. J. Respir. Cell Mol. Biol. 2007, 36, 8–12. [Google Scholar]
- Reshmi, G.; Pillai, M.R. Beyond HPV: oncomirs as new players in cervical cancer. FEBS Lett. 2008, 582, 4113–4116. [Google Scholar] [CrossRef]
- Bala, S.; Marcos, M.; Szabo, G. Emerging role of microRNAs in liver diseases. World J. Gastroenterol. 2009, 15, 5633–5640. [Google Scholar] [CrossRef]
- Esau, C.; Kang, X.; Peralta, E.; Hanson, E.; Marcusson, E.G.; Ravichandran, L.V.; Sun, Y.; Koo, S.; Perera, R.J.; Jain, R.; Dean, N.M.; Freier, S.M.; Bennett, C.F.; Lollo, B.; Griffey, R. MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 2004, 279, 52361–52365. [Google Scholar]
- Krützfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with 'antagomirs'. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef]
- Weiler, J.; Hunziker, J.; Hall, J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2006, 13, 496–502. [Google Scholar] [CrossRef]
- Stenvang, J.; Kauppinen, S. MicroRNAs as targets for antisense-based therapeutics. Expert Opin. Biol. Ther. 2008, 8, 59–81. [Google Scholar] [CrossRef]
- Mattes, J.; Collison, A.; Foster, P.S. Emerging role of microRNAs in disease pathogenesis and strategies for therapeutic modulation. Curr. Opin. Mol. Ther. 2008, 10, 150–157. [Google Scholar]
- Petri, A.; Lindow, M.; Kauppinen, S. MicroRNA silencing in primates: towards development of novel therapeutics. Cancer Res. 2009, 69, 393–395. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Umbach, J.L.; Cullen, B.R. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009, 23, 1151–1164. [Google Scholar] [CrossRef]
- Umbach, J.L.; Kramer, M.F.; Jurak, I.; Karnowski, H.W.; Coen, D.M.; Cullen, B.R. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 2008, 454, 780–783. [Google Scholar]
- Nachmani, D.; Stern-Ginossar, N.; Sarid, R.; Mandelboim, O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 2009, 5, 376–385. [Google Scholar] [CrossRef]
- Nachmani, D.; Lankry, D.; Wolf, D.G.; Mandelboim, O. The human cytomegalovirus microRNA miR-UL112 acts synergistically with a cellular microRNA to escape immune elimination. Nat. Immunol. 2010, 11, 806–813. [Google Scholar]
- Moens, U. Silencing viral microRNA as a novel antiviral therapy? J. Biomed. Biotechnol. 2009. Article ID 419539. [Google Scholar]
- He, S.; Yang, Z.; Skogerbo, G.; Ren, F.; Cui, H.; Zhao, H.; Chen, R.; Zhao, Y. The properties and functions of virus encoded microRNA, siRNA, and other small noncoding RNA. Crit. Rev. Microbiol. 2008, 34, 175–188. [Google Scholar] [CrossRef]
- Chang, J.; Guo, J.T.; Jiang, D.; Guo, H.; Taylor, J.M.; Block, T.M. Liver-specific microRNA miR-122 enhances the replication of hepatitis C virus in nonhepatic cells. J. Virol. 2008, 82, 8215–8223. [Google Scholar]
- Jopling, C.L.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005, 309, 1577–1581. [Google Scholar] [CrossRef]
- Henke, J.I.; Goergen, D.; Zheng, J.; Song, Y.; Schuttler, C.G.; Fehr, C.; Junemann, C.; Niepmann, M. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008, 27, 3300–3310. [Google Scholar]
- Randall, G.; Panis, M.; Cooper, J.D.; Tellinghuisen, T.L.; Sukhodolets, K.E.; Pfeffer, S.; Landthaler, M.; Landgraf, P.; Kan, S.; Lindenbach, B.D.; Chien, M.; Weir, D.B.; Russo, J.J.; Ju, J.; Brownstein, M.J.; Sheridan, R.; Sander, C.; Zavolan, M.; Tuschl, T.; Rice, C.M. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. USA 2007, 104, 12884–12889. [Google Scholar]
- Elmen, J.; Lindow, M.; Silahtaroglu, A.; Bak, M.; Christensen, M.; Lind-Thomsen, A.; Hedtjarn, M.; Hansen, J.B.; Hansen, H.F.; Straarup, E.M.; McCullagh, K.; Kearney, P.; Kauppinen, S. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucl. Acid. Res. 2008, 36, 1153–1162. [Google Scholar]
- Elmen, J.; Lindow, M.; Schutz, S.; Lawrence, M.; Petri, A.; Obad, S.; Lindholm, M.; Hedtjarn, M.; Hansen, H.F.; Berger, U.; Gullans, S.; Kearney, P.; Sarnow, P.; Straarup, E.M.; Kauppinen, S. LNA-mediated microRNA silencing in non-human primates. Nature 2008, 452, 896–899. [Google Scholar]
- Lanford, R.E.; Hildebrandt-Eriksen, E.S.; Petri, A.; Persson, R.; Lindow, M.; Munk, M.E.; Kauppinen, S.; Orum, H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 2010, 327, 198–201. [Google Scholar] [CrossRef]
- Hammond, S.M.; Bernstein, E.; Beach, D.; Hannon, G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000, 404, 293–296. [Google Scholar] [CrossRef]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef]
- Paddison, P.J.; Caudy, A.A.; Bernstein, E.; Hannon, G.J.; Conklin, D.S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002, 16, 948–958. [Google Scholar] [CrossRef]
- Brummelkamp, T.R.; Bernards, R.; Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Science 2002, 296, 550–553. [Google Scholar]
- Amarzguioui, M.; Rossi, J.J.; Kim, D. Approaches for chemically synthesized siRNA and vector-mediated RNAi. FEBS Lett. 2005, 579, 5974–5981. [Google Scholar] [CrossRef]
- Song, E.; Lee, S.K.; Wang, J.; Ince, N.; Ouyang, N.; Min, J.; Chen, J.; Shankar, P.; Lieberman, J. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat. Med. 2003, 9, 347–351. [Google Scholar]
- Bumcrot, D.; Manoharan, M.; Koteliansky, V.; Sah, D.W. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat. Chem. Biol. 2006, 2, 711–719. [Google Scholar] [CrossRef]
- Kretschmer-Kazemi, F.R.; Sczakiel, G. The activity of siRNA in mammalian cells is related to structural target accessibility: a comparison with antisense oligonucleotides. Nucl. Acid. Res. 2003, 31, 4417–4424. [Google Scholar] [CrossRef]
- Overhoff, M.; Alken, M.; Far, R.K.; Lemaitre, M.; Lebleu, B.; Sczakiel, G.; Robbins, I. Local RNA target structure influences siRNA efficacy: a systematic global analysis. J. Mol. Biol. 2005, 348, 871–881. [Google Scholar]
- Jackson, A.L.; Linsley, P.S. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov. 2010, 9, 57–67. [Google Scholar]
- Amarzguioui, M.; Lundberg, P.; Cantin, E.; Hagstrom, J.; Behlke, M.A.; Rossi, J.J. Rational design and in vitro and in vivo delivery of Dicer substrate siRNA. Nat. Protoc. 2006, 1, 508–517. [Google Scholar]
- Muhonen, P.; Holthofer, H. Bioinformatic approaches to siRNA selection and optimization. Methods Mol. Biol. 2010, 623, 93–107. [Google Scholar] [CrossRef]
- Mittal, V. Improving the efficiency of RNA interference in mammals. Nat. Rev. Genet. 2004, 5, 355–365. [Google Scholar]
- Moschos, S.A.; Spinks, K.; Williams, A.E.; Lindsay, M.A. Targeting the lung using siRNA and antisense based oligonucleotides. Curr. Pharm. Des. 2008, 14, 3620–3627. [Google Scholar]
- Bitko, V.; Musiyenko, A.; Shulyayeva, O.; Barik, S. Inhibition of respiratory viruses by nasally administered siRNA. Nat. Med. 2005, 11, 50–55. [Google Scholar] [CrossRef]
- DeVincenzo, J.; Cehelsky, J.E.; Alvarez, R.; Elbashir, S.; Harborth, J.; Toudjarska, I.; Nechev, L.; Murugaiah, V.; Van Vliet, A.; Vaishnaw, A.K.; Meyers, R. Evaluation of the safety, tolerability and pharmacokinetics of ALN-RSV01, a novel RNAi antiviral therapeutic directed against respiratory syncytial virus (RSV). Antivir. Res. 2008, 77, 225–231. [Google Scholar]
- Das, A.T.; Brummelkamp, T.R.; Westerhout, E.M.; Vink, M.; Madiredjo, M.; Bernards, R.; Berkhout, B. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J. Virol. 2004, 78, 2601–2605. [Google Scholar] [CrossRef]
- Westerhout, E.M.; Ooms, M.; Vink, M.; Das, A.T.; Berkhout, B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucl. Acid. Res. 2005, 33, 796–804. [Google Scholar]
- ter Brake, O.; 't Hooft, K.; Liu, Y.P.; Centlivre, M.; von Eije, K.J.; Berkhout, B. Lentiviral vector design for multiple shRNA expression and durable HIV-1 inhibition. Mol. Ther. 2008, 16, 557–564. [Google Scholar] [CrossRef]
- Geisbert, T.W.; Lee, A.C.; Robbins, M.; Geisbert, J.B.; Honko, A.N.; Sood, V.; Johnson, J.C.; de Jong, S.; Tavakoli, I.; Judge, A.; Hensley, L.E.; Maclachlan, I. Postexposure protection of non-human primates against a lethal Ebola virus challenge with RNA interference: a proof-of-concept study. Lancet 2010, 375, 1896–1905. [Google Scholar]
- Morrissey, D.V.; Lockridge, J.A.; Shaw, L.; Blanchard, K.; Jensen, K.; Breen, W.; Hartsough, K.; Machemer, L.; Radka, S.; Jadhav, V.; Vaish, N.; Zinnen, S.; Vargeese, C.; Bowman, K.; Shaffer, C.S.; Jeffs, L.B.; Judge, A.; Maclachlan, I.; Polisky, B. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 2005, 23, 1002–1007. [Google Scholar]
- Rossi, J.J. RNAi therapeutics: SNALPing siRNAs in vivo. Gene Ther. 2006, 13, 583–584. [Google Scholar] [CrossRef]
- Nowak, M.; Wyszko, E.; Fedoruk-Wyszomirska, A.; Pospieszny, H.; Barciszewska, M.Z.; Barciszewski, J. A new and efficient method for inhibition of RNA viruses by DNA interference. FEBS J. 2009, 276, 4372–4380. [Google Scholar]
- Sullenger, B.A.; Gallardo, H.F.; Ungers, G.E.; Gilboa, E. Overexpression of TAR sequences renders cells resistant to human immunodeficiency virus replication. Cell 1990, 63, 601–608. [Google Scholar] [CrossRef]
- Sullenger, B.A.; Gallardo, H.F.; Ungers, G.E.; Gilboa, E. Analysis of trans-acting response decoy RNA-mediated inhibition of human immunodeficiency virus type 1 transactivation. J. Virol. 1991, 65, 6811–6816. [Google Scholar]
- Bohjanen, P.R.; Colvin, R.A.; Puttaraju, M.; Been, M.D.; Garcia-Blanco, M.A. A small circular TAR RNA decoy specifically inhibits Tat-activated HIV-1 transcription. Nucl. Acid. Res. 1996, 24, 3733–3738. [Google Scholar] [CrossRef]
- Anderson, J.; Li, M.J.; Palmer, B.; Remling, L.; Li, S.; Yam, P.; Yee, J.K.; Rossi, J.; Zaia, J.; Akkina, R. Safety and efficacy of a lentiviral vector containing three anti-HIV genes (CCR5 ribozyme, tat-rev siRNA, and TAR decoy) in SCID-hu mouse-derived T cells. Mol. Ther. 2007, 15, 1182–1188. [Google Scholar]
- Anderson, J.S.; Javien, J.; Nolta, J.A.; Bauer, G. Preintegration HIV-1 inhibition by a combination lentiviral vector containing a chimeric TRIM5 alpha protein, a CCR5 shRNA, and a TAR decoy. Mol. Ther. 2009, 17, 2103–2114. [Google Scholar] [CrossRef]
- Bahner, I.; Kearns, K.; Hao, Q.L.; Smogorzewska, E.M.; Kohn, D.B. Transduction of human CD34+ hematopoietic progenitor cells by a retroviral vector expressing an RRE decoy inhibits human immunodeficiency virus type 1 replication in myelomonocytic cells produced in long-term culture. J. Virol. 1996, 70, 4352–4360. [Google Scholar]
- Kohn, D.B.; Bauer, G.; Rice, C.R.; Rothschild, J.C.; Carbonaro, D.A.; Valdez, P.; Hao, Q.; Zhou, C.; Bahner, I.; Kearns, K.; Brody, K.; Fox, S.; Haden, E.; Wilson, K.; Salata, C.; Dolan, C.; Wetter, C.; Aguilar-Cordova, E.; Church, J. A clinical trial of retroviral-mediated transfer of a rev-responsive element decoy gene into CD34(+) cells from the bone marrow of human immunodeficiency virus-1-infected children. Blood 1999, 94, 368–371. [Google Scholar]
- Wyatt, J.R.; Vickers, T.A.; Roberson, J.L.; Buckheit, R.W., Jr.; Klimkait, T.; DeBaets, E.; Davis, P.W.; Rayner, B.; Imbach, J.L.; Ecker, D.J. Combinatorially selected guanosine-quartet structure is a potent inhibitor of human immunodeficiency virus envelope-mediated cell fusion. Proc. Natl. Acad. Sci. USA 1994, 91, 1356–1360. [Google Scholar]
- Pinskaya, M.; Romanova, E.; Volkov, E.; Deprez, E.; Leh, H.; Brochon, J.C.; Mouscadet, J.F.; Gottikh, M. HIV-1 integrase complexes with DNA dissociate in the presence of short oligonucleotides conjugated to acridine. Biochemistry 2004, 43, 8735–8743. [Google Scholar]
- Mescalchin, A.; Wünsche, W.; Laufer, S.D.; Grohmann, D.; Restle, T.; Sczakiel, G. Specific binding of a hexanucleotide to HIV-1 reverse transcriptase: a novel class of bioactive molecules. Nucl. Acid. Res. 2006, 34, 5631–5637. [Google Scholar] [CrossRef]
- Mescalchin, A.; Wünsche, W.; Sczakiel, G. Specific recognition of proteins by array-bound hexanucleotides. Angew. Chem. Int. Ed. Engl. 2011. In Press. [Google Scholar]
- Bunka, D.H.; Stockley, P.G. Aptamers come of age - at last. Nat. Rev. Microbiol. 2006, 4, 588–596. [Google Scholar] [CrossRef]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef]
- Robertson, D.L.; Joyce, G.F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 1990, 344, 467–468. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Morris, K.N.; Jensen, K.B.; Julin, C.M.; Weil, M.; Gold, L. High affinity ligands from in vitro selection: complex targets. Proc. Natl. Acad. Sci. USA 1998, 95, 2902–2907. [Google Scholar]
- Patel, D.J.; Suri, A.K. Structure, recognition and discrimination in RNA aptamer complexes with cofactors, amino acids, drugs and aminoglycoside antibiotics. J. Biotechnol. 2000, 74, 39–60. [Google Scholar]
- Hermann, T.; Patel, D.J. Adaptive recognition by nucleic acid aptamers. Science 2000, 287, 820–825. [Google Scholar] [CrossRef]
- Niazi, J.H.; Lee, S.J.; Gu, M.B. Single-stranded DNA aptamers specific for antibiotics tetracyclines. Bioorg. Med. Chem. 2008, 16, 7245–7253. [Google Scholar] [CrossRef]
- Dausse, E.; Da Rocha, G.S.; Toulme, J.J. Aptamers: a new class of oligonucleotides in the drug discovery pipeline? Curr. Opin. Pharmacol. 2009, 9, 602–607. [Google Scholar] [CrossRef]
- Jayasena, S.D. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar]
- Cerchia, L.; Hamm, J.; Libri, D.; Tavitian, B.; de Franciscis, V. Nucleic acid aptamers in cancer medicine. FEBS Lett. 2002, 528, 12–16. [Google Scholar]
- Proske, D.; Blank, M.; Buhmann, R.; Resch, A. Aptamers--basic research, drug development, and clinical application. Appl. Microbiol. Biotechnol. 2005, 69, 367–374. [Google Scholar]
- Famulok, M. Allosteric aptamers and aptazymes as probes for screening approaches. Curr. Opin. Mol. Ther. 2005, 7, 137–143. [Google Scholar]
- Que-Gewirth, N.S.; Sullenger, B.A. Gene therapy progress and prospects: RNA aptamers. Gene Ther. 2007, 14, 283–291. [Google Scholar] [CrossRef]
- Ulrich, H.; Trujillo, C.A.; Nery, A.A.; Alves, J.M.; Majumder, P.; Resende, R.R.; Martins, A.H. DNA and RNA aptamers: from tools for basic research towards therapeutic applications. Comb. Chem. High Throughput. Screen. 2006, 9, 619–632. [Google Scholar] [CrossRef]
- Gold, L. RNA as the catalyst for drug screening. Nat. Biotechnol. 2002, 20, 671–672. [Google Scholar] [CrossRef]
- McNamara, J.O.; Andrechek, E.R.; Wang, Y.; Viles, K.D.; Rempel, R.E.; Gilboa, E.; Sullenger, B.A.; Giangrande, P.H. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006, 24, 1005–1015. [Google Scholar] [CrossRef]
- Ng, E.W.; Shima, D.T.; Calias, P.; Cunningham, E.T., Jr.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar]
- Srisawat, C.; Engelke, D.R. Selection of RNA aptamers that bind HIV-1 LTR DNA duplexes: strand invaders. Nucl. Acid. Res. 2010, 38, 8306–8315. [Google Scholar] [CrossRef]
- Duconge, F.; Toulme, J.J. In vitro selection identifies key determinants for loop-loop interactions: RNA aptamers selective for the TAR RNA element of HIV-1. RNA 1999, 5, 1605–1614. [Google Scholar] [CrossRef]
- Boiziau, C.; Dausse, E.; Yurchenko, L.; Toulme, J.J. DNA aptamers selected against the HIV-1 trans-activation-responsive RNA element form RNA-DNA kissing complexes. J. Biol. Chem. 1999, 274, 12730–12737. [Google Scholar]
- Sekkai, D.; Dausse, E.; Di Primo, C.; Darfeuille, F.; Boiziau, C.; Toulme, J.J. In vitro selection of DNA aptamers against the HIV-1 TAR RNA hairpin. Antisense Nucleic Acid Drug Dev. 2002, 12, 265–274. [Google Scholar] [CrossRef]
- Kolb, G.; Reigadas, S.; Castanotto, D.; Faure, A.; Ventura, M.; Rossi, J.J.; Toulme, J.J. Endogenous expression of an anti-TAR aptamer reduces HIV-1 replication. RNA Biol. 2006, 3, 150–156. [Google Scholar]
- Watrin, M.; Von Pelchrzim, F.; Dausse, E.; Schroeder, R.; Toulme, J.J. In vitro selection of RNA aptamers derived from a genomic human library against the TAR RNA element of HIV-1. Biochemistry 2009, 48, 6278–6284. [Google Scholar]
- Kikuchi, K.; Umehara, T.; Fukuda, K.; Kuno, A.; Hasegawa, T.; Nishikawa, S. A hepatitis C virus (HCV) internal ribosome entry site (IRES) domain III-IV-targeted aptamer inhibits translation by binding to an apical loop of domain IIId. Nucl. Acid. Res. 2005, 33, 683–692. [Google Scholar] [CrossRef]
- Kikuchi, K.; Umehara, T.; Nishikawa, F.; Fukuda, K.; Hasegawa, T.; Nishikawa, S. Increased inhibitory ability of conjugated RNA aptamers against the HCV IRES. Biochem. Biophys. Res. Commun. 2009, 386, 118–123. [Google Scholar]
- Konno, K.; Fujita, S.; Iizuka, M.; Nishikawa, S.; Hasegawa, T.; Fukuda, K. Isolation and characterization of RNA aptamers specific for the HCV minus-IRES domain I. Nucl. Acid. Symp. Ser. 2008, 52, 493–494. [Google Scholar]
- Schneider, D.J.; Feigon, J.; Hostomsky, Z.; Gold, L. High-affinity ssDNA inhibitors of the reverse transcriptase of type 1 human immunodeficiency virus. Biochemistry 1995, 34, 9599–9610. [Google Scholar] [CrossRef]
- Burke, D.H.; Scates, L.; Andrews, K.; Gold, L. Bent pseudoknots and novel RNA inhibitors of type 1 human immunodeficiency virus (HIV-1) reverse transcriptase. J. Mol. Biol. 1996, 264, 650–666. [Google Scholar]
- Andreola, M.L.; Pileur, F.; Calmels, C.; Ventura, M.; Tarrago-Litvak, L.; Toulme, J.J.; Litvak, S. DNA aptamers selected against the HIV-1 RNase H display in vitro antiviral activity. Biochemistry 2001, 40, 10087–10094. [Google Scholar]
- Joshi, P.; Prasad, V.R. Potent inhibition of human immunodeficiency virus type 1 replication by template analog reverse transcriptase inhibitors derived by SELEX (systematic evolution of ligands by exponential enrichment). J. Virol. 2002, 76, 6545–6557. [Google Scholar] [CrossRef]
- Hannoush, R.N.; Min, K.L.; Damha, M.J. Diversity-oriented solid-phase synthesis and biological evaluation of oligonucleotide hairpins as HIV-1 RT RNase H inhibitors. Nucl. Acid. Res. 2004, 32, 6164–6175. [Google Scholar] [CrossRef]
- Zhang, Z.; Blank, M.; Schluesener, H.J. Nucleic acid aptamers in human viral disease. Arch. Immunol. Ther. Exp. 2004, 52, 307–315. [Google Scholar]
- Toulmé, J.J.; Di Primo, C.; Boucard, D. Regulating eukaryotic gene expression with aptamers. FEBS Lett. 2004, 567, 55–62. [Google Scholar]
- DeStefano, J.J.; Cristofaro, J.V. Selection of primer-template sequences that bind human immunodeficiency virus reverse transcriptase with high affinity. Nucl. Acid. Res. 2006, 34, 130–139. [Google Scholar] [CrossRef]
- Tarrago-Litvak, L.; Andreola, M.L.; Nevinsky, G.A.; Sarih-Cottin, L.; Litvak, S. The reverse transcriptase of HIV-1: from enzymology to therapeutic intervention. FASEB J. 1994, 8, 497–503. [Google Scholar]
- Tuerk, C.; MacDougal, S.; Gold, L. RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase. Proc. Natl. Acad. Sci. USA 1992, 89, 6988–6992. [Google Scholar]
- Jaeger, J.; Restle, T.; Steitz, T.A. The structure of HIV-1 reverse transcriptase complexed with an RNA pseudoknot inhibitor. EMBO J. 1998, 17, 4535–4542. [Google Scholar] [CrossRef]
- Kensch, O.; Connolly, B.A.; Steinhoff, H.J.; McGregor, A.; Goody, R.S.; Restle, T. HIV-1 reverse transcriptase-pseudoknot RNA aptamer interaction has a binding affinity in the low picomolar range coupled with high specificity. J. Biol. Chem. 2000, 275, 18271–18278. [Google Scholar]
- Chaloin, L.; Lehmann, M.J.; Sczakiel, G.; Restle, T. Endogenous expression of a high-affinity pseudoknot RNA aptamer suppresses replication of HIV-1. Nucl. Acid. Res. 2002, 30, 4001–4008. [Google Scholar]
- Held, D.M.; Kissel, J.D.; Saran, D.; Michalowski, D.; Burke, D.H. Differential susceptibility of HIV-1 reverse transcriptase to inhibition by RNA aptamers in enzymatic reactions monitoring specific steps during genome replication. J. Biol. Chem. 2006, 281, 25712–25722. [Google Scholar]
- Mayer, G. The chemical biology of aptamers. Angew. Chem. Int. Ed. Engl. 2009, 48, 2672–2689. [Google Scholar] [CrossRef]
- White, R.; Rusconi, C.; Scardino, E.; Wolberg, A.; Lawson, J.; Hoffman, M.; Sullenger, B. Generation of species cross-reactive aptamers using ´toggle´ SELEX. Mol. Ther. 2001, 4, 567–573. [Google Scholar] [CrossRef]
- Padilla, R.; Sousa, R. A Y639F/H784A T7 RNA polymerase double mutant displays superior properties for synthesizing RNAs with non-canonical NTPs. Nucl. Acid. Res. 2002, 30, e138. [Google Scholar] [CrossRef]
- Chelliserrykattil, J.; Ellington, A.D. Evolution of a T7 RNA polymerase variant that transcribes 2'-O-methyl RNA. Nat. Biotechnol. 2004, 22, 1155–1160. [Google Scholar] [CrossRef]
- Keefe, A.D.; Cload, S.T. SELEX with modified nucleotides. Curr. Opin. Chem. Biol. 2008, 12, 448–456. [Google Scholar] [CrossRef]
- Beigelman, L.; McSwiggen, J.A.; Draper, K.G.; Gonzalez, C.; Jensen, K.; Karpeisky, A.M.; Modak, A.S.; Matulic-Adamic, J.; DiRenzo, A.B.; Haeberli, P. Chemical modification of hammerhead ribozymes. Catalytic activity and nuclease resistance. J. Biol. Chem. 1995, 270, 25702–25708. [Google Scholar]
- Cox, J.C.; Rudolph, P.; Ellington, A.D. Automated RNA selection. Biotechnol. Prog. 1998, 14, 845–850. [Google Scholar] [CrossRef]
- Cox, J.C.; Hayhurst, A.; Hesselberth, J.; Bayer, T.S.; Georgiou, G.; Ellington, A.D. Automated selection of aptamers against protein targets translated in vitro: From gene to aptamer. Nucl. Acid. Res. 2002, 30, e108. [Google Scholar]
- Vater, A.; Klussmann, S. Toward third-generation aptamers: Spiegelmers and their therapeutic prospects. Curr. Opin. Drug Discov. Dev. 2003, 6, 253–261. [Google Scholar]
- Ryan, S.M.; Mantovani, G.; Wang, X.; Haddleton, D.M.; Brayden, D.J. Advances in PEGylation of important biotech molecules: delivery aspects. Expert Opin. Drug Deliv. 2008, 5, 371–383. [Google Scholar]
- Jackson, A.L.; Linsley, P.S. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov. 2010, 9, 57–67. [Google Scholar]
- Laufer, S.D.; Restle, T. Peptide-mediated cellular delivery of oligonucleotide-based therapeutics in vitro: Quantitative evaluation of overall efficacy employing easy to handle reporter systems. Curr. Pharm. Des. 2008, 14, 3637–3655. [Google Scholar] [CrossRef]
- Whitehead, K.A.; Langer, R.; Anderson, D.G. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 2009, 8, 129–138. [Google Scholar]
- Laufer, S.D.; Detzer, A.; Sczakiel, G.; Restle, T. Selected Strategies for the Delivery of siRNA In vitro and In vivo. In RNA Technologies and Their Applications; Erdmann, V.A., Barciszewski, J., Eds.; Springer-Verlag: Berlin Heidelberg, Germany, 2010; pp. 29–58. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Mescalchin, A.; Restle, T. Oligomeric Nucleic Acids as Antivirals. Molecules 2011, 16, 1271-1296. https://doi.org/10.3390/molecules16021271
Mescalchin A, Restle T. Oligomeric Nucleic Acids as Antivirals. Molecules. 2011; 16(2):1271-1296. https://doi.org/10.3390/molecules16021271
Chicago/Turabian StyleMescalchin, Alessandra, and Tobias Restle. 2011. "Oligomeric Nucleic Acids as Antivirals" Molecules 16, no. 2: 1271-1296. https://doi.org/10.3390/molecules16021271
APA StyleMescalchin, A., & Restle, T. (2011). Oligomeric Nucleic Acids as Antivirals. Molecules, 16(2), 1271-1296. https://doi.org/10.3390/molecules16021271