Inhibition of 11b-HSD1 by Tetracyclic Triterpenoids from Euphorbia kansui
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of Compounds
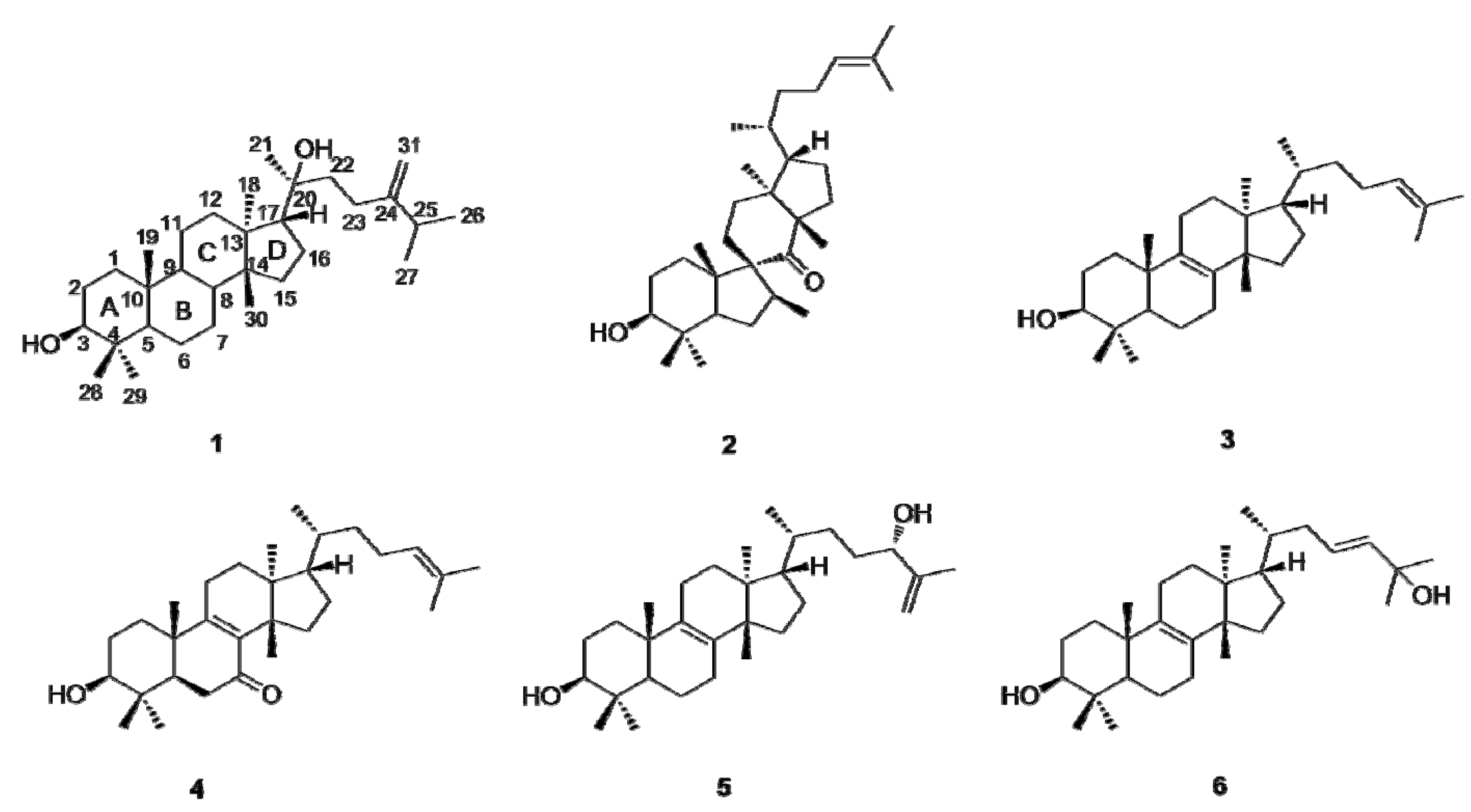
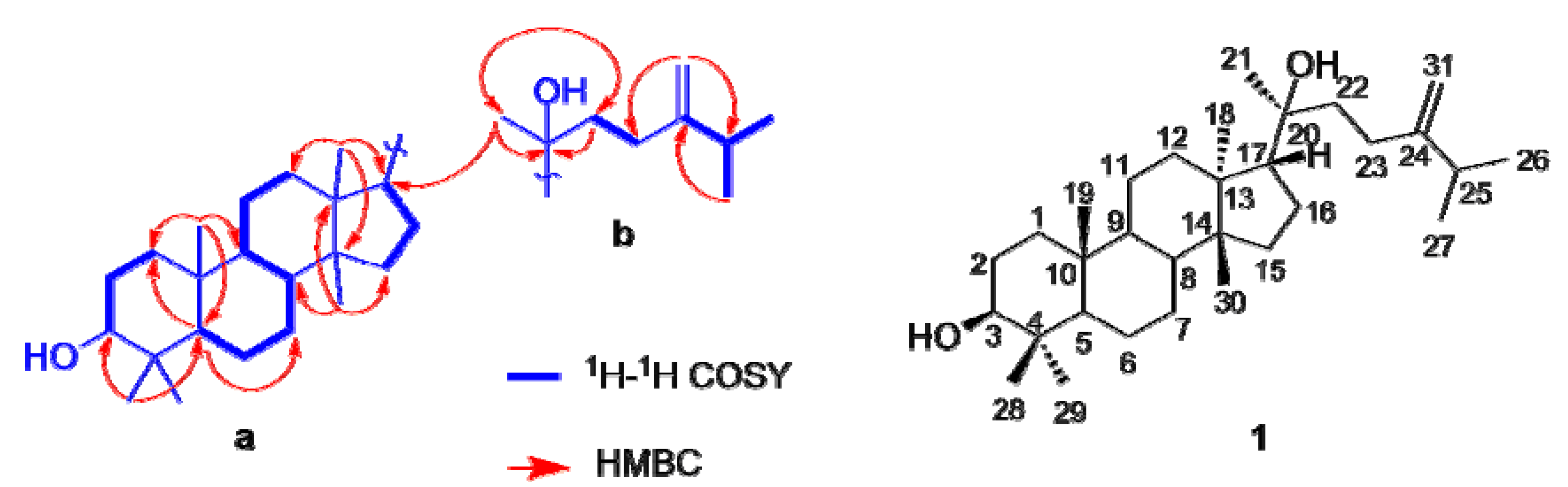
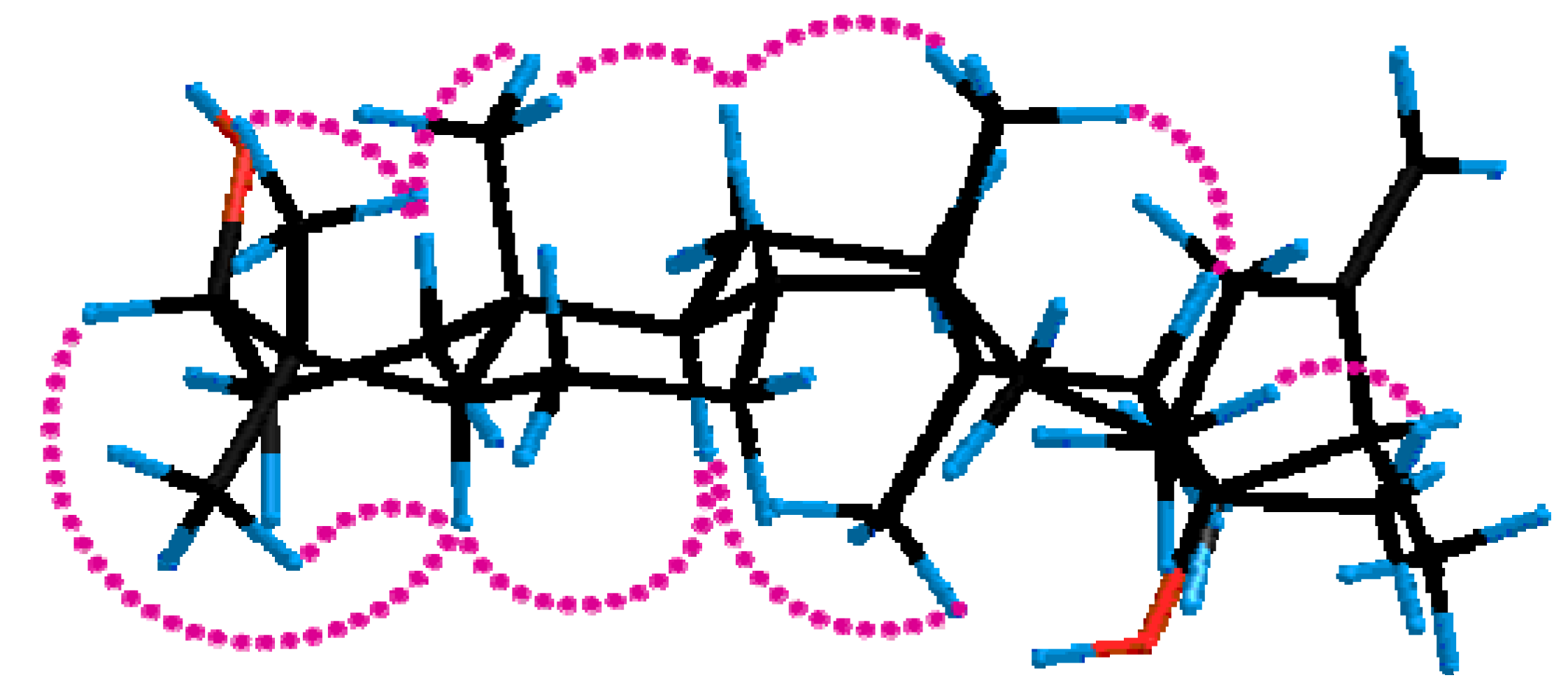
2.2. Inhibition of 11β-HSD1
| Compounds | Mouse 11β-HSD1 (IC50) | Mouse 11β-HSD2 (IC50) | Mouse HSD2/HSD1 | Human 11β-HSD1 (IC50) | Human 11β-HSD2 (IC50) | Human HSD2/HSD1 |
|---|---|---|---|---|---|---|
| Compound 1 | 78.44 nM | >1 mM | >12748 | 34.86 nM | 8.179 μM | 234.6 |
| Compound 2 | 1.077 μM | >1 mM | >928 | 1.115 μM | 2.626 μM | 2.35 |
| Compound 3 | 80.52 nM | >1 mM | >12419 | 16.08 nM | 0.3952 μM | 24.6 |
| Compound 4 | 13.36 nM | >1 mM | >74850 | 2.815 nM | 0.107 μM | 38 |
| Compound 5 | 49.46 nM | >1 mM | >20218 | 26.47 nM | 1.687 μM | 63.7 |
| Compound 6 | 294.7 nM | >1 mM | >3393 | 15.99 nM | 0.6664 μM | 41.7 |
2.3. In Silico Study of the Activities of Compounds

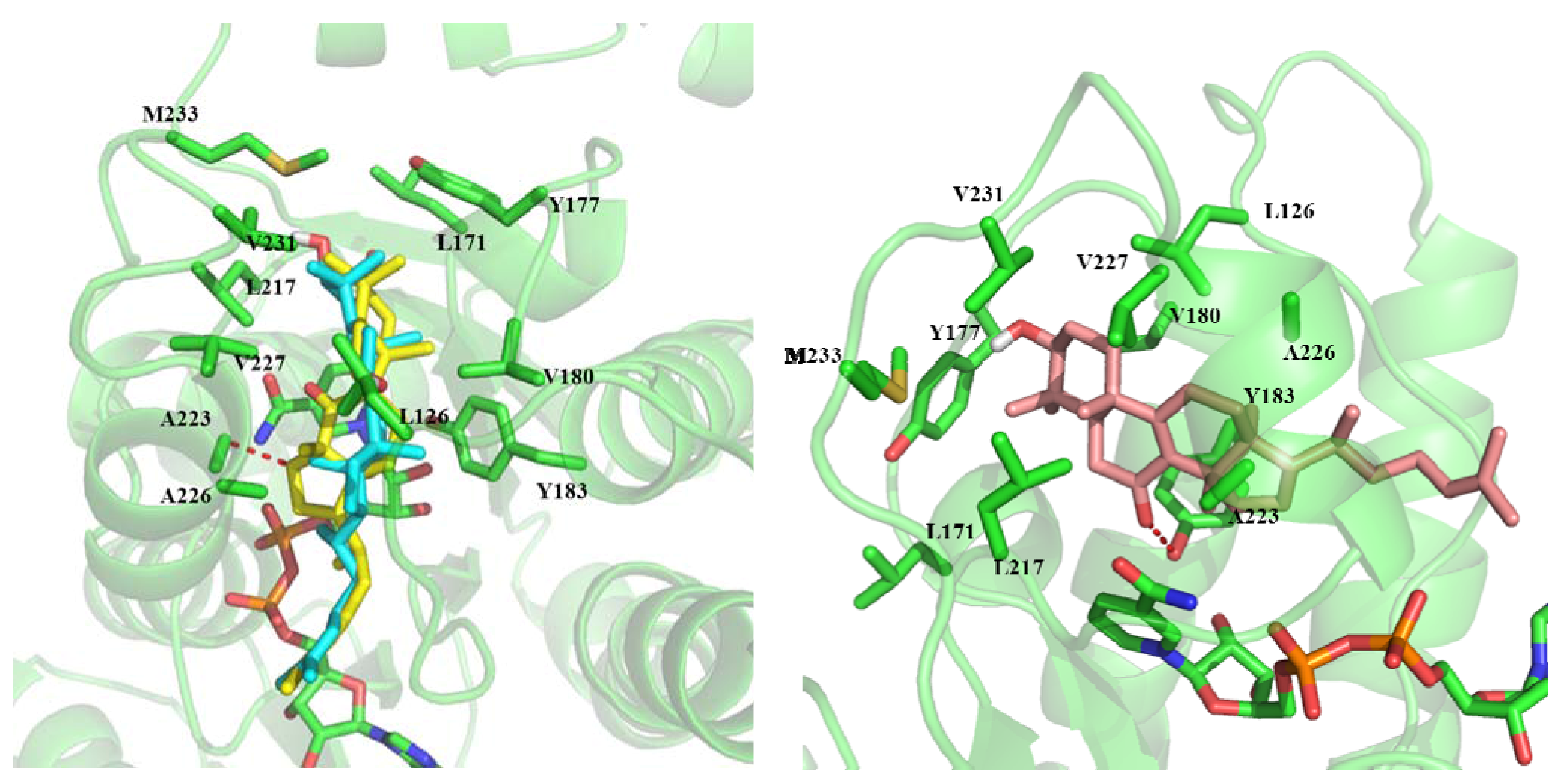

3. Experimental
3.1. General
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Computational Methods
3.5. Biological Testing of Compounds
4. Conclusions
Supplementary Materials
Acknowledgments
Conflict of Interest
- Sample Availability: Contact the authors.
References
- Stumvoll, M.; Goldstein, B.J.; van Haeften, T.W. Type 2 diabetes: Principles of pathogenesis and therapy. Lancet 2005, 365, 1333–1346. [Google Scholar] [CrossRef]
- Yang, W.Y.; Lu, J.M.; Weng, J.P.; Jia, W.P.; Ji, L.N.; Xiao, J.Z.; Shan, Z.Y.; Liu, J.; Tian, H.M.; Ji, Q.H.; et al. Prevalence of Diabetes among Men and Women in China. N. Engl. J. Med. 2010, 12, 1090–1101. [Google Scholar]
- Evans, F.J.; Taylor, S.E. Pro-inflammatory, Tumour-promoting and anti-tumour diterpenes of the plant families Euphorbiaceae and Thymelaeaceae. Prog. Chem. Org. Nat. Prod. 1983, 44, 1–99. [Google Scholar]
- Günther, G.; Martinek, T.; Dombi, G.; Hohmann, J.; Vasas, A. Structural characterisation and dynamic NMR studies of a new peracylated macrocyclic diterpene. Magn. Reson. Chem. 1999, 37, 365–370. [Google Scholar]
- Appendino, G.; Belloro, E.; Tron, G.C.; Jakupovic, J.; Ballero, M. Diterpenoids from Euphorbia pithyusa subsp. cupanii. J. Nat. Prod. 1999, 10, 1399–1404. [Google Scholar]
- Vogg, G.; Mattes, E.; Rothenburger, J.; Hertkorn, N.; Achatz, S.; Sanderman, H. Tumor promoting diterpenes from Euphorbia leuconeura L. Phytochemistry 1999, 2, 289–295. [Google Scholar]
- He, W.D.; Cik, M.; Lesage, A.; Linden, I.V.D.; Kinpe, N.D.; Appendino, G.; Bracke, J.; Mathenge, S.G.; Mudida, F.P.; Leysen, J.E.; et al. Kirkinine, A New Daphnane Orthoester with Potent Neurotrophic Activity from Synaptolepis kirkii. J. Nat. Prod. 2000, 9, 1185–1187. [Google Scholar]
- Amit, R.; Shailendra, S. Limonoids: Overview of Significant Bioactive Triterpenes Distributed in Plants Kingdom. Biol. Pharm. Bull. 2006, 2, 191–201. [Google Scholar]
- Guo, J.; Fang, X.; Di, Y.T.; Hua, H.M.; Hao, X.J. Kansuinine J, A new macrocyclic diterpenoid from the roots of Euphorbia kansui. Chin. Chem. Lett. 2010, 8, 943–946. [Google Scholar]
- Halaweish, F.T.; Kronberg, S.; Rice, J.A. Rodent and Ruminant Ingestive Response to Flavonoids in Euphorbia esula. J. Chem. Ecol. 2002, 5, 1073–1082. [Google Scholar]
- Liu, L.G.; Meng, J.C.; Tan, R.X. New macrocyclic diterpenoids from Euphorbia esula. Planta Med. 2002, 3, 244–248. [Google Scholar]
- Akihisa, T.; Kithsiri Wijeratne, E.M.; Tokuda, H.; Enjo, F.; Toriumi, M.; Kimura, Y.; Koike, K.; Nikaido, T.; Tezuka, Y.; Nishino, H. Eupha-7,9(11),24-trien-3a-ol (“Antiquol C”) and Other Triterpenes from Euphorbia antiquorum Latex and Their Inhibitory Effects on Epstein-Barr Virus Activation. J. Nat. Prod. 2002, 65, 158–162. [Google Scholar] [CrossRef]
- Jiang, Z.H.; Tanaka, T.; Hirata, H.; Fukuoka, R.; Kouno, I. The first euphane-type triterpene tridesmosides and bisdesmoside from Rhoiptelea chiliantha. Tetrahedron 1997, 50, 16999–17008. [Google Scholar]
- Akihisa, T.; Kimura, Y.; Kokke, W.C.M.C.; Takase, S.; Yasukawa, K.; Tamura, T. Tirucalla-5,24-dien-3β-ol[(13α,14β,17α,20S)-lanosta-5,24-dien-3β-ol] and three other Δ5-unsaturated tirucallanes from the roots of Bryonia dioica Jacq.: The first naturally occurring C-10 methylated tetracyclic triterpene alcohols with a Δ5-monounsaturated skeleton. J. Chem. Soc. Perk. Trans. 1996, 19, 2379–2384. [Google Scholar]
- Arai, Y.; Hirohara, M.; Ageta, H. Fern constituents: Three new skeletal triterpenoid hydrocarbons isolated from Polypodiodes niponica. Tetrahedron Lett. 1989, 30, 7209–7212. [Google Scholar] [CrossRef]
- Abe, I.; Rohmer, M. Enzymic cyclization of 2,3-dihydrosqualene and squalene 2,3-epoxide by squalene cyclases: From pentacyclic to tetracyclic triterpenes. J. Chem. Soc. Perk. Trans. 1994, 7, 783–791. [Google Scholar]
- Mamta, M.; Yogendra, N.; Sushil, K. Euphane triterpenoid and lipid constituents from Butea monosperma. Phytochemistry 2000, 54, 835–838. [Google Scholar] [CrossRef]
- Ge, R.; Huang, Y.; Liang, G.; Li, X. 11b-hydroxysteroids dehydrogenase type 1 inhibitors as promising therapeutic drugs for diabetes: Status and development. Curr. Med. Chem. 2010, 5, 412–422. [Google Scholar]
- Chen, Q.; Li, Y.D.; Li, K.L. Clinical study on the effect of Tangwei Dressing applied to acupoint on gastrointestinal hormones of patients with diabetic gastric paresis. Chin. Tradit. Patent Med. 2011, 33, 208–211. [Google Scholar]
- Gao, J.M.; Bosco, D.A.; Powers, E.T.; Kelly, J.W. Localized thermodynamic coupling between hydrogen bonding and microenvironment polarity substantially stabilizes proteins. Nat. Struct. Mol. Biol. 2009, 16, 684–690. [Google Scholar] [CrossRef]
- Guo, J.; He, H.P.; Fang, X.; Di, Y.T.; Li, S.L.; Zhang, Z.; Leng, Y.; Hua, H.M.; Hao, X.J. Kansuinone, A novel triterpene with an unusual skeleton from Euphorbia kansui. Tetrahedron Lett. 2010, 48, 6286–6289. [Google Scholar]
- Cao, D.; Su, Y.L.; Yang, J.S. Triterpene Constitutents From Euphorbia nematocypha HAND-MAZZ. Acta Pharm. Sin. 1992, 6, 445–451. [Google Scholar]
- Wang, L.Y.; Wang, N.L.; Yao, X.S.; Syohei, M.; Susumu, K. Euphane and Tirucallane Triterpenes from the Roots of Euphorbia kansui and Their in Vitro Effects on the Cell Division of Xenopus. J. Nat. Prod. 2003, 66, 630–633. [Google Scholar] [CrossRef]
- Leong, Y.W.; Harrison, L.J. (20R, 23E)-Eupha-8,23-diene-3,25-diol from Tripetalum cymosum. Phytochemistry 1999, 5, 849–857. [Google Scholar]
- Ttott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comp. Chem. 2010, 31, 455–461. [Google Scholar]
- DeLano, W.L. The PyMOL Molecular Graphics System. Available online: http://www.pymol.org (accessed on 14 August 2009).
- Stewart, P.M.; Draper, N. 11β-Hydroxysteroid dehydrogenase and the pre-receptor regulation of corticosteroid hormone action. J. Endocrinol. 2005, 186, 251–271. [Google Scholar] [CrossRef]
- Seckl, J.R.; Walker, B.R. Minireview: 11β-hydroxysteroid dehydrogenase type 1—A tissue-specific amplifier of glucocorticoid action. Endocrinology 2001, 142, 1371–1376. [Google Scholar] [CrossRef]
- Yang, H.Y.; Dou, W.; Lou, J.; Leng, Y.; Shen, J.H. Discovery of novel inhibitors of 11beta-hydroxysteroid dehydrogenase type 1 by docking and pharmacophore modeling. Bioorg. Med. Chem. Lett. 2008, 18, 1340–1345. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Guo, J.; Zhou, L.-Y.; He, H.-P.; Leng, Y.; Yang, Z.; Hao, X.-J. Inhibition of 11b-HSD1 by Tetracyclic Triterpenoids from Euphorbia kansui. Molecules 2012, 17, 11826-11838. https://doi.org/10.3390/molecules171011826
Guo J, Zhou L-Y, He H-P, Leng Y, Yang Z, Hao X-J. Inhibition of 11b-HSD1 by Tetracyclic Triterpenoids from Euphorbia kansui. Molecules. 2012; 17(10):11826-11838. https://doi.org/10.3390/molecules171011826
Chicago/Turabian StyleGuo, Jie, Li-Yan Zhou, Hong-Ping He, Ying Leng, Zhen Yang, and Xiao-Jiang Hao. 2012. "Inhibition of 11b-HSD1 by Tetracyclic Triterpenoids from Euphorbia kansui" Molecules 17, no. 10: 11826-11838. https://doi.org/10.3390/molecules171011826




