MiR-15a Decreases Bovine Mammary Epithelial Cell Viability and Lactation and Regulates Growth Hormone Receptor Expression
Abstract
:1. Introduction
2. Results and Discussion
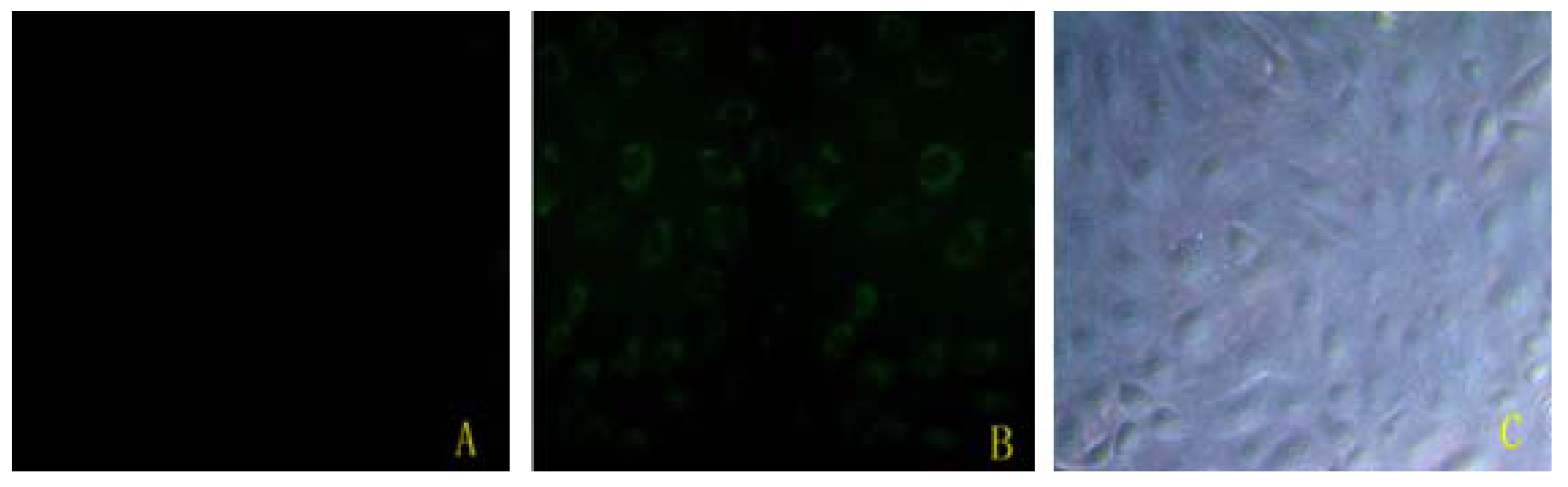
2.1. Bta-miR-15a Was Transfected Efficiently
2.2. Ghr Were Predicted to Be the Target of Bta-miR-15a
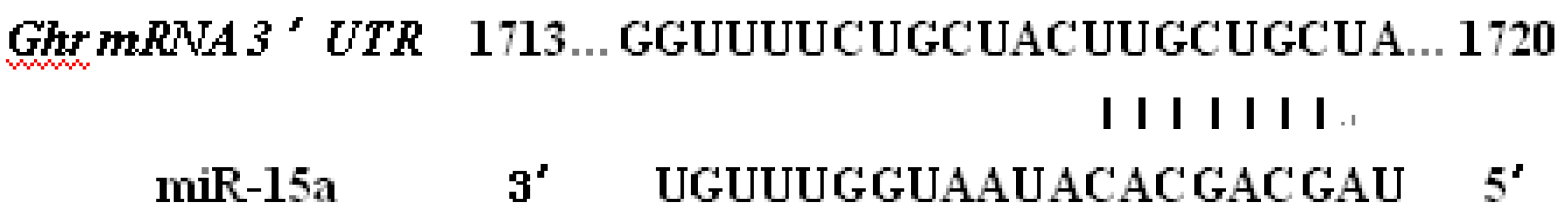
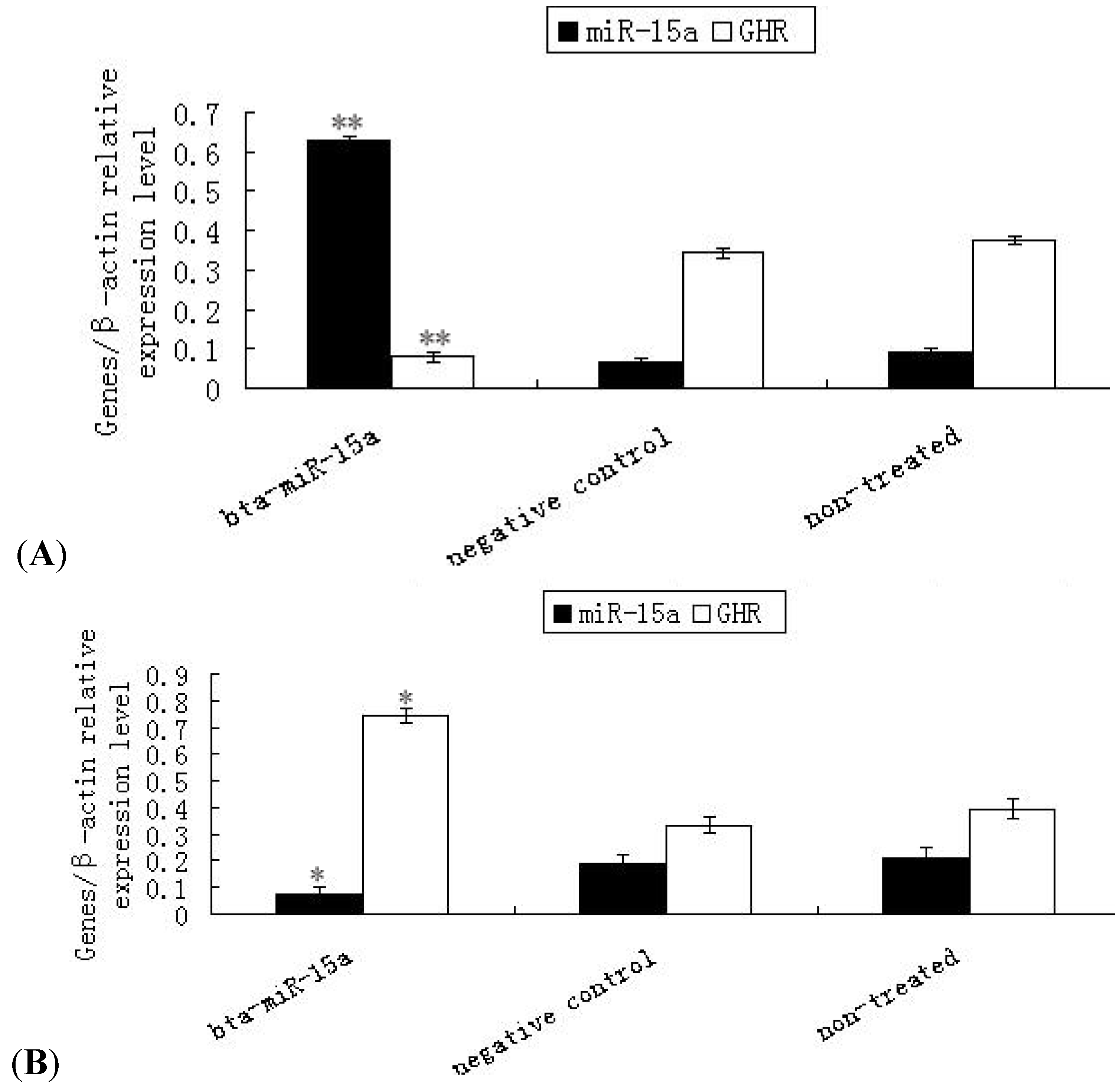
2.3. Expression of Ghr mRNA in Bta-miR-15a Mimics/Inhibitor Transfected BMECs
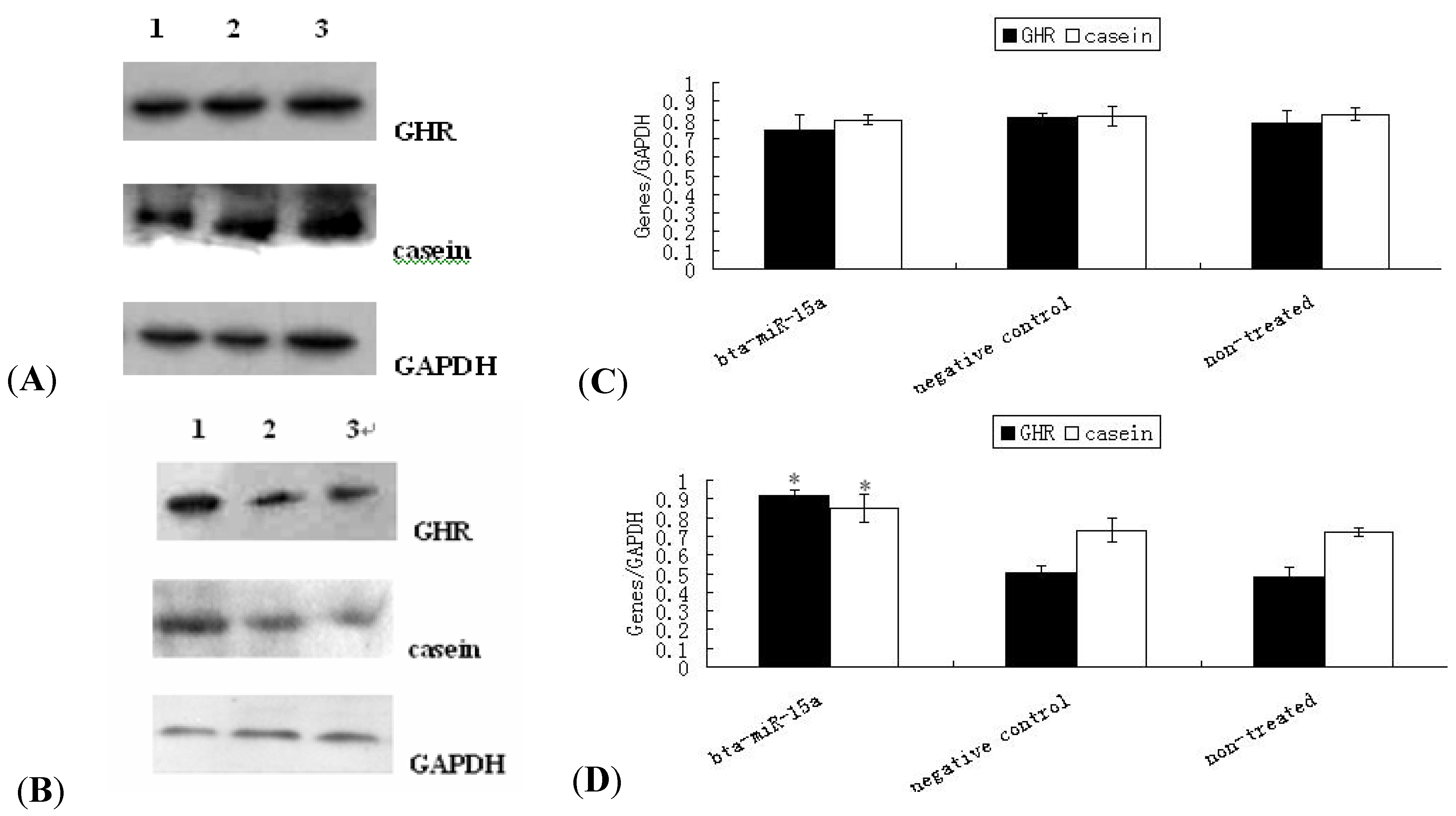
2.4. Expression of GHR Protein and Casein Synthesis in Bta-miR-15a Mimics/Inhibitor Transfected BMECs
2.5. Bta-miR-15a Reduced Cell Viability
| A. Effect of bta-miR-15a mimics transfection on BMECs viability. | |
| Group | Viability (%) |
| miR-15a mimics 30 nM | 64.69 ± 0.60 b |
| Negative control | 83.52 ± 1.26 a |
| Non-transfected group | 82.35 ± 0.80 a |
| B. Effect of bta-miR-15a inhibitor transfection on BMECs viability | |
| Group | Viability (%) |
| miR-15a inhibitor 30nM | 89.43 ± 0.18 b |
| Negative control | 82.78 ± 0.62 a |
| Non-transfected group | 83.15 ± 0.50 a |
2.6. Discussion
3. Experimental
3.1. Cell Culture
3.2. Transient Transfection
3.3. Quantitative Real-Time PCR Analysis
3.4. Western Blotting
3.5. Quantification of Cell Viability
3.6. Statistical Analysis
4. Conclusions
Acknowledgments
References
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Gregory, R.I.; Chendrimada, T.P.; Cooch, N.; Shiekhattar, R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005, 123, 631–640. [Google Scholar] [CrossRef]
- Doench, J.G.; Sharp, P.A. Specificity of microRNA target selection in translational repression. Genes Dev. 2004, 18, 504–511. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Olsen, P.H.; Ambros, V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 1999, 216, 671–680. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. MicroRNA as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Chen, J.F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006, 38, 228–233. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasqulnelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef]
- Ambros, V. MicroRNA pathways in flies and worms: Growth, Death, Fat, Stress, and Timing. Cell 2003, 113, 673–676. [Google Scholar] [CrossRef]
- Wang, C.; Li, Q. Identification of differentially expressed microRNAs during the development of Chinese Murine mammary gland. J. Genet. Genomics 2007, 34, 966–973. [Google Scholar] [CrossRef]
- Bionaz, M.; Loor, J.J. Identification of reference genes for quantitative real-time PCR in the bovine mammary gland during the lactation cycle. Physiol. Genomics 2007, 29, 312–319. [Google Scholar] [CrossRef]
- Gu, Z.; Eleswarapu, S.; Jiang, H. Identification and characterization of microRNAs from the bovine adipose tissue and mammary gland. FEBS Lett. 2007, 581, 981–988. [Google Scholar] [CrossRef]
- Bonci, D.; Coppola, V.; Musumeci, M.; Addario, A.; Giuffrida, R.; Memeo, L.; D’Urso, L.; Pagliuca, A.; Biffoni, M.; Labbaye, C.; et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat. Med. 2008, 14, 1271–1277. [Google Scholar] [CrossRef]
- Bandi, N.; Zbinden, S.; Gugger, M.; Amold, M.; Kocher, V.; Hasan, L.; Kappeler, A.; Brunner, T.; Vassella, E. MiR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009, 69, 5553–5559. [Google Scholar] [CrossRef]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. MiR-15 and miR-16 induce apoptosis by targeting Bcl-2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar]
- Ageilan, R.I.; Calin, G.A.; Croce, C.M. MiR-15a and miR-16-1 in cancer: Discovery, Function and future perspectives. Cell Death Differ. 2010, 17, 215–220. [Google Scholar] [CrossRef]
- Zhou, Y.; Akers, R.M.; Jiang, H. Growth hormone can induce expression of four major milk protein genes in transfected MAC-T cells. J. Dairy Sci. 2008, 91, 100–108. [Google Scholar] [CrossRef]
- Akers, R.M. Major advances associated with hormone and growth factor regulation of mammary growth and lactation in dairy cows. J. Dairy Sci. 2006, 89, 1222–1234. [Google Scholar] [CrossRef]
- Goffin, V.; Kelly, P.A. The Prolactin/Growth Hormone Receptor Family: Structure/Function Relationships. J. Mammary Gland Biol. Neoplasia 1997, 2, 7–17. [Google Scholar] [CrossRef]
- Zych, S.; Szatkowska, I.; Czerniawska-Piatkowska, E. Growth hormone signaling pathways. Postepy Biochem. 2006, 52, 367–372. [Google Scholar]
- Kleinberg, D.L.; Ruan, W. IGF-I, GH, and Sex steroid effects in normal mammary gland development. J. Mammary Gland Biol. Neoplasia 2008, 13, 353–360. [Google Scholar] [CrossRef]
- Kamiński, S.; Malewski, T.; Ahman, A.; Wójcik, E.; Ruść, A.; Oleński, K.; Jakubczak, A.; Sazanov, A.A. Towards an integrated approach to study SNPs and expression of candidate genes associated with milk protein biosynthesis. Genetika 2008, 44, 532–538. [Google Scholar]
- Rahmatalla, S.A.; Müller, U.; Strucken, E.M.; Reissmann, M.; Brockmann, G.A. The F279Y polymorphism of the GHR gene and its relation to milk production and somatic cell score in German Holstein dairy cattle. J. Appl. Genet. 2011, 52, 459–465. [Google Scholar] [CrossRef]
- Tanaka, T.; Haneda, S.; Imakawa, K.; Sakai, S.; Nagaoka, K. A microRNA, miR-101a, Controls mammary gland development by regulating cyclooxygenase-2 expression. Differentiation 2009, 77, 181–187. [Google Scholar] [CrossRef]
- Bhattacharya, R.; Nicoloso, M.; Arvizo, R.; Wang, E.; Cortez, A.; Rossi, S.; Calin, G.A.; Mukherjee, P. MiR-15a and MiR-16 control Bmi-1 expression in ovarian cancer. Cancer Res. 2009, 69, 9090–9095. [Google Scholar]
- Sakamoto, K.; Komatsu, T.; Kobayashi, T.; Rose, M.T.; Aso, H.; Hagino, A.; Obara, Y. Growth hormone acts on the synthesis and secretion of alpha-casein in bovine mammary epithelial cells. J. Dairy Res. 2005, 72, 264–270. [Google Scholar] [CrossRef]
- Sakamoto, K.; Yano, T.; Kobayashi, T.; Hagino, A.; Aso, H.; Obara, Y. Growth hormone suppresses the expression of IGFBP-5, And promotes the IGF-I-induced phosphorylation of Akt in bovine mammary epithelial cells. Domest. Anim. Endocrin. 2007, 32, 260–272. [Google Scholar] [CrossRef]
- Mao, J.; Molenaar, A.J.; Wheeler, T.T.; Seyfert, H.M. STAT5 binding contributes to lactational stimulation of promoter III expressing the bovine acetyl-CoA carboxylase alpha-encoding gene in the mammary gland. J. Mol. Endocrinol. 2002, 29, 73–88. [Google Scholar] [CrossRef]
- Gallego, M.I.; Binart, N.; Robinson, G.W.; Okagaki, R.; Coschigano, K.T.; Perry, J.; Kopchick, J.J.; Oka, T.; Kelly, P.A.; Hennighausen, L. Prolactin, Growth Hormone, and Epidermal Growth Factor Activate Stat5 in Different Compartments of Mammary Tissue and Exert Different and overlapping Developmental Effects. Dev. Biol. 2001, 229, 63–175. [Google Scholar]
- Boutinaud, M.; Guinard-Flamenta, J.; Jammes, H. The number and activity of mammary epithelial cells, Determining factors for milk production. Reprod. Nutr. Dev. 2004, 44, 499–508. [Google Scholar] [CrossRef]
- Boutinaud, M.; Lollivier, V.; Finot, L.; Bruckmaier, R.M.; Lacasse, P. Mammary cell activity and turnover in dairy cows treated with the prolactin-release inhibitor quinagolide and milked once daily. J. Dairy Sci. 2011, 95, 177–187. [Google Scholar]
- Gorski, D.H.; Leal, A.J. Inhibition of endothelial cell activation by the homeobox gene Gax. J. Surg. Res. 2003, 111, 91–99. [Google Scholar] [CrossRef]
- Alcorn, J.; Lu, X.; Moscow, J.A.; McNamara, P.J. Transporter gene expression in Lactating and nonlactating human mammary epithelial cells using real-time reverse transcription polymerase chain reaction. J. Pharmacol. Exp. Ther. 2002, 303, 487–496. [Google Scholar] [CrossRef]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ibarra, I.; Erlich, Y.; Senthil, K.; Muthuswamy, S.K.; Sachiidanandam, R.; Hannon, G.J. A role for microRNAs in maintenance of mouse mammary epithelial. Genes Dev. 2007, 21, 3238–3243. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the bovine mammary gland epithelial cells (BMECs) are available from the authors.
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Li, H.-M.; Wang, C.-M.; Li, Q.-Z.; Gao, X.-J. MiR-15a Decreases Bovine Mammary Epithelial Cell Viability and Lactation and Regulates Growth Hormone Receptor Expression. Molecules 2012, 17, 12037-12048. https://doi.org/10.3390/molecules171012037
Li H-M, Wang C-M, Li Q-Z, Gao X-J. MiR-15a Decreases Bovine Mammary Epithelial Cell Viability and Lactation and Regulates Growth Hormone Receptor Expression. Molecules. 2012; 17(10):12037-12048. https://doi.org/10.3390/molecules171012037
Chicago/Turabian StyleLi, Hui-Ming, Chun-Mei Wang, Qing-Zhang Li, and Xue-Jun Gao. 2012. "MiR-15a Decreases Bovine Mammary Epithelial Cell Viability and Lactation and Regulates Growth Hormone Receptor Expression" Molecules 17, no. 10: 12037-12048. https://doi.org/10.3390/molecules171012037
APA StyleLi, H.-M., Wang, C.-M., Li, Q.-Z., & Gao, X.-J. (2012). MiR-15a Decreases Bovine Mammary Epithelial Cell Viability and Lactation and Regulates Growth Hormone Receptor Expression. Molecules, 17(10), 12037-12048. https://doi.org/10.3390/molecules171012037




