Experimental Methodologies and Evaluations of Computer-Aided Drug Design Methodologies Applied to a Series of 2-Aminothiophene Derivatives with Antifungal Activities
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis and Antifungal Activity

| Comp. | n | Ar | MIC (μg/mL) | |
|---|---|---|---|---|
| C. neoformans ICB59 | Candida krusei LM13 | |||
| 1 | 1 | Ph | 2,500 | 1,250 |
| 2 | 1 | 4-NO2-Ph | 1,250 | 2,500 |
| 3 | 1 | 2-Thiophenyl | 156 | >10,000 |
| 4 | 1 | 2-Indole | 1,250 | >10,000 |
| 5 | 1 | 4-F-Ph | 2,500 | >10,000 |
| 6 | 1 | 4-Br-Ph | 2,500 | >10,000 |
| 7 | 1 | 4-Morpholine | 312 | >10,000 |
| 8 | 1 | 4-OEt-Ph | 2,500 | >10,000 |
| 9 | 1 | 4-Cl-Ph | 5,000 | 2,500 |
| 10 | 1 | 4-OMe-Ph | 1,250 | 2,500 |
| 11 | 1 | 2,3-Cl-Ph | 156 | >10,000 |
| 12 | 1 | 3,4-Cl-Ph | >10,000 | 5,000 |
| 13 | 1 | 2,4-Cl-Ph | 78 | 78 |
| 14 | 1 | 3,4,5-OMe-Ph | 156 | 78 |
| 15 | 1 | 2-OMe,5-Br-Ph | 156 | 78 |
| 16 | 1 | 8-Quinoline | 156 | 5,000 |
| 17 | 2 | 4-NO2-Ph | 1,250 | 128 |
| 18 | 2 | 2-Indole | 78 | >10,000 |
| 19 | 2 | 4-OBz-Ph | >10,000 | 625 |
| 20 | 2 | 4-Br-Ph | >10,000 | >10,000 |
| 21 | 2 | 4-Cl-Ph | >10,000 | >10,000 |
| 22 | 2 | 4-OMe-Ph | 156 | 1,250 |
| 23 | 2 | 4-F-Ph | 5,000 | 625 |
| 24 | 2 | 2,3-Cl-Ph | 156 | 156 |
| 25 | 2 | 3,4-Cl-Ph | 1,250 | 312 |
| 26 | 2 | 2,4-Cl-Ph | 2,500 | >10,000 |
| 27 | 2 | 3,4,5-OMe-Ph | 156 | 2,500 |
| 28 | 2 | 2-OMe,5-Br-Ph | 10,000 | 10,000 |
| 29 | 2 | 4-Pyrrolidine-Ph | 2,500 | 2,500 |
| 30 | 2 | 3-Thiophenyl | 2,500 | >10,000 |
| 31 | 2 | 3,4-OBz-Ph | 2,500 | 1,250 |
| 32 | 2 | 8-Quinoline | 78 | 625 |
| 33 | 2 | 4-Morpholine-Ph | 2,500 | 2,500 |
| 34 | 3 | 4-Me-Ph | 156 | 2,500 |
| 35 | 3 | 4-iPr-Ph | 1,250 | 2,500 |
| 36 | 3 | Ph | >10,000 | 2,500 |
| 37 | 3 | 4-NO2-Ph | 10,000 | 625 |
| 38 | 3 | 2-indole | 312 | 2,500 |
| 39 | 3 | 4-OBz-Ph | 10,000 | >10,000 |
| 40 | 3 | 4-Br-Ph | 2,500 | 5,000 |
| 41 | 3 | 4-Cl-Ph | 1,250 | 2,500 |
| 42 | 3 | 4-OMe-Ph | 312 | 2,500 |
| 43 | 3 | 4-OEt-Ph | 5,000 | 2,500 |
| 44 | 3 | 3,4-Cl-Ph | 1,250 | 2,500 |
| 45 | 3 | 2,4-Cl-Ph | >10,000 | 2,500 |
| 46 | 3 | 3,4,5-OMe-Ph | 1,250 | 1,250 |
| 47 | 3 | 2-OMe,5-Br-Ph | 10,000 | 5,000 |
| 48 | 3 | 3-Thiophenyl | 2,500 | >10,000 |
| 49 | 3 | 8-Quinoline | 78 | 625 |
| 50 | 3 | 4-Et-Ph | 2,500 | 2,500 |
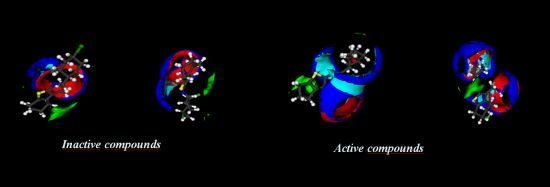
2.2. Electronic Surfaces

2.3. Chemometric Studies Using the Program VolSurf+
2.3.1. CPCA (Consensus PCA)
| PC | % explained variance from original data |
|---|---|
| 1 | 41.26 |
| 2 | 22.69 |
| 3 | 3.06 |
| 4 | 1.05 |
| 5 | 0.00 |
| PC | % explained variance from original data |
|---|---|
| 1 | 45.03 |
| 2 | 20.11 |
| 3 | 4.00 |
| 4 | 0.98 |
| 5 | 0.00 |

| PC | % explained variance from original data |
|---|---|
| 1 | 43.77 |
| 2 | 20.58 |
| 3 | 2.33 |
| 4 | 1.71 |
| 5 | 0.09 |
2.3.2. PCA
| PC | % explained variance from original data |
|---|---|
| 1 | 39.80 |
| 2 | 22.75 |
| 3 | 3.56 |
| 4 | 1.00 |
| 5 | 0.08 |

- SOLY-intrinsic solubility,
- LOGS10 and 11-intrinsic solubilities calculated at different pHs (pH = 10 and pH = 11),
- CACO2 and SKIN-permeability/solubility in skin cells and CACO2,
- L1 and 2 GS-solubility for Legendre-polynomials that are used to differentiate solubility between similar compounds,
- FLEX_RB–A parameter related to flexibility and the number of rotational links. The Flex descriptor represents the maximum flexibility of a molecule. It is the result of the average of the differences between the maximum and minimum distance of every atom with the others searched on 50 random conformers). Flex_RB descriptor is the ratio between Flex and the number of rotable bonds.
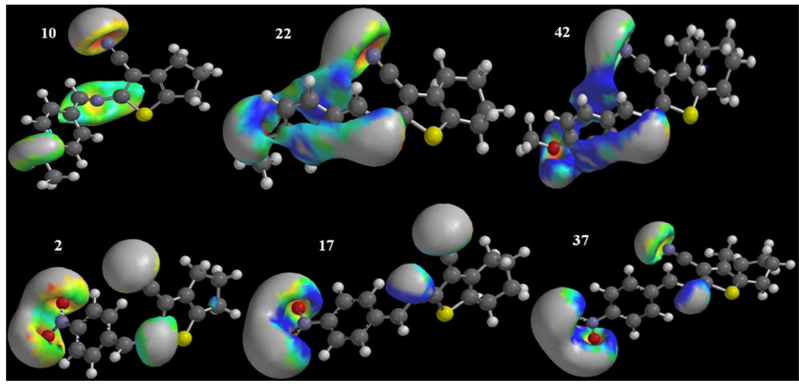
2.4. QSAR-3D and PLS
- H7m–related to the probability of interaction between atoms and topological distance 7.
- R4u+, R7u+ and R3v+–related to the maximum steric contributions to the molecule shape related to the atoms with topological distance of 4, 7 and 3, respectively. These descriptor have greater values for higher influence on the shape and the lower geometric distance between them.
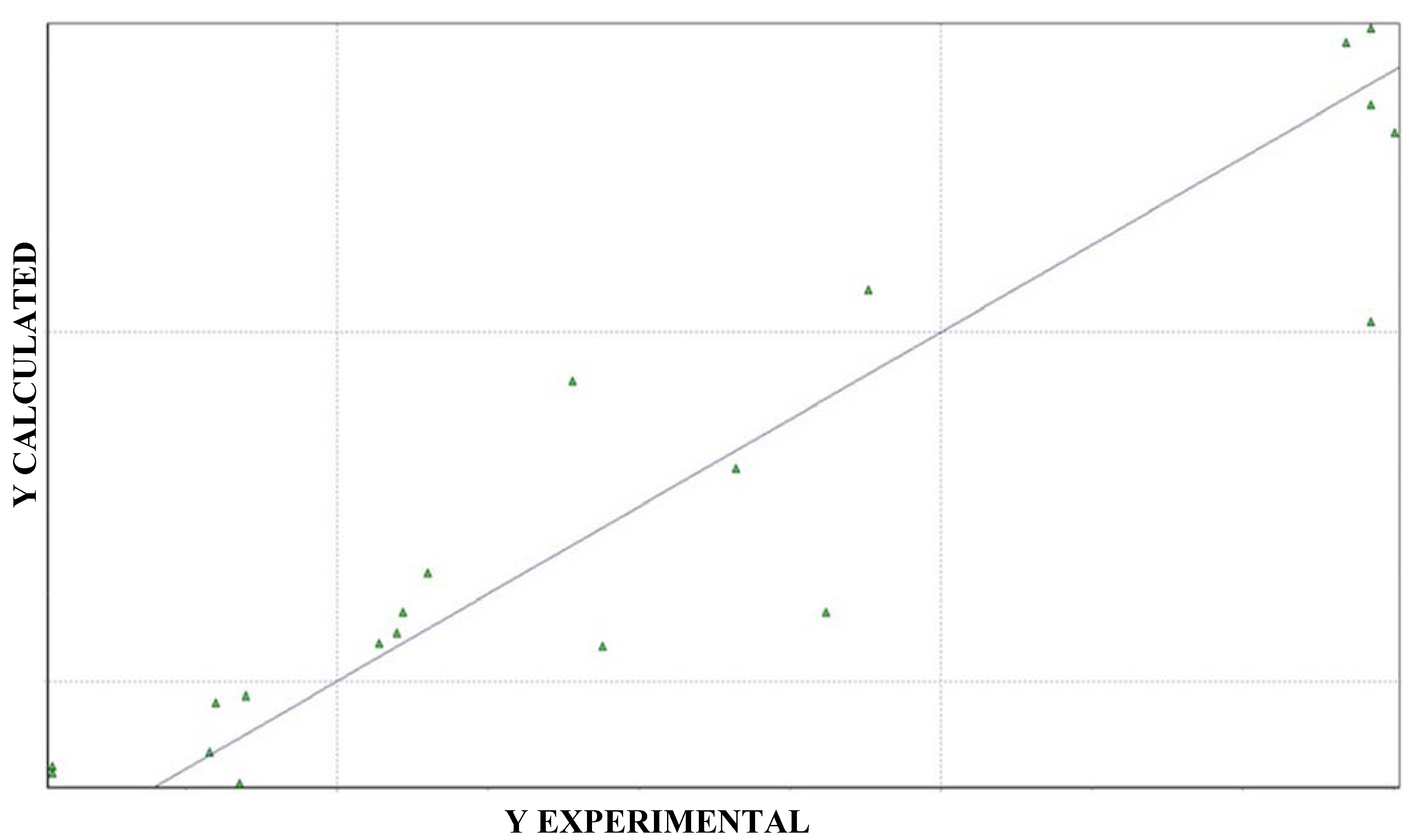
| pIC50 calculated | pIC50 observed | Residues | SD |
|---|---|---|---|
| 2.54 | 2.38 | 0.16 | 0.26 |
| 3.87 | 3.70 | 0.17 | |
| 3.72 | 3.67 | 0.05 | |
| 2.28 | 1.53 | 0.75 | |
| 2.27 | 2.09 | 0.18 |
| LV | SDEC | SDEP | R2 | Q2 |
|---|---|---|---|---|
| 5 | 0.18 | 0.48 | 0.91 | 0.77 |

3. Experimental
3.1. Antifungal Activity
3.2. Molecular Modelling
3.3. Chemometrics
3.4. QSAR
3.5. PLS
4. Conclusions
Conflict of Interest
Acknowledgements
- Sample Availability: Samples of the compounds 1–50 are available from the authors.
References and Notes
- Brown, J.M. Fungal infections in bone marrow transplant patients. Curr. Opin. Infect. Dis. 2004, 17, 347–352. [Google Scholar] [CrossRef]
- Bergold, A.M.; Georgiadis, S. New antifungic drugs: A review. Visão Acadêmica 2004, 5, 159–172. [Google Scholar]
- Ram, V.J.; Goel, A.; Shukla, P.K.; Kapil, A. Synthesis of thiophenes and thieno[3,2-c]pyran-4-ones as antileishmanial and antifungal agents. Bioorg. Med. Chem. Lett. 1997, 7, 3101–3106. [Google Scholar] [CrossRef]
- Chobot, V.; Buchta, V.; Jahodárová, H.; Pour, M.; Opletal, L.; Jahodár, L.; Harant, P. Antifungal activity of a thiophenepolyine from Leuzeacarthamoides. Phytotherapy 2003, 74, 288–290. [Google Scholar]
- Wardakhan, W.W.; Louca, N.A.; Kamel, M.M. The reaction of 2 Aminocyclohexeno[b]thiophene derivatives with ethoxycarbonylisothiocyanate: Synthesis of fused thiophene derivatives with antibacterial and antifungal activities. Acta Chim. Slov. 2007, 54, 229–241. [Google Scholar]
- Shaaban, M.R.; Saleh, T.S.; Farag, A.M. Synthesis and antimicrobial evaluation of new thiophene and 1,3,4-thiadiazole derivatives. 2009, 78, 151–159. [Google Scholar]
- Islor, A.M.; Kalluraya, B.; Pai, K.S. Synthesis, characterization and biological activities of some new benzo[b]thiophene derivatives. Eur. J. Med. Chem. 2010, 45, 825–830. [Google Scholar] [CrossRef]
- Fokialakis, N.; Cantrell, C.L.; Duke, S.O.; Skaltsounis, A.L.; Wedge, D.E. Antifungal activity of thiophenes from Echinops ritro. J. Agric. Food Chem. 2006, 54, 1651–1655. [Google Scholar] [CrossRef]
- Pinto, E.; Queiroz, M.J.R.P.; Vale-Silva, L.A.; Oliveira, J.F.; Begoin, A.; Begoin, J.M.; Kirsch, G. Antifungal activity of synthetic di(hetero)arylamines based on the benzo[b]thiophene moiety. Bioorg. Med. Chem. 2008, 16, 8172–8177. [Google Scholar] [CrossRef]
- Mendonça Junior, F.J.B.; Lima-Neto, R.G.; de Oliveira, T.B.; Lima, M.C.A.; Pitta, I.R.; Galdino, S.L.; da Cruz, R.M.D.; de Araújo, R.S.A.; Neves, R.P. Synthesis and evaluation of the antifungal activity of 2-(substituted-amino)-4,5-dialkyl-thiophene-3-carbonitrile derivatives. Lat. Am. J. Pharm. 2011, 30, 1492–1499. [Google Scholar]
- Abi-Said, D.; Anaissie, E.; Uzum, O.; Raad, I.; Pinzcowski, H.; Vartivarian, S. The epidemiology of hematogenouscandidiasis caused by different Candida species. Clin. Infect. Dis. 1997, 24, 1122–1128. [Google Scholar]
- Berrouane, Y.F.; Hollis, R.J.; Pfaller, M.A. Strain variation among and antifungal susceptibilities of isolates of Candida krusei. J. Clin. Microbiol. 1996, 34, 1856–1858. [Google Scholar]
- Pfaller, M.A.; Diekema, D.J.; Gibbs, D.L.; Newell, V.A.; Nagy, E.; Dobiasova, S.; Rinaldi, M.; Barton, R.; Veselov, A. Global Antifungal Surveillance Group. Candida krusei, a multidrug-resistant opportunistic fungal pathogen: Geographic and temporal trends from the ARTEMIS DISK antifungal surveillance program, 2001 to 2005. J. Clin. Microbiol. 2008, 46, 515–521. [Google Scholar]
- Merz, W.G.; Karp, J.E.; Schron, D.; Saral, R. Increased incidence of fungemia caused by candida krusei. J. Clin. Microbiol. 1986, 24, 581–584. [Google Scholar]
- Cheung, M.C.; Rachlis, A.R.; Shumak, S.L. A cryptic cause of cryptococcal meningitis. Can. Med. Assoc. J. 2003, 168, 451–452. [Google Scholar]
- Wormley, F.L., Jr.; Perfect, J.R.; Steele, C.; Cox, G.M. Protection against Cryptococcosis by Using a Murine Gamma Interferon-Producing Cryptococcus neoformans Strain. Infect. Immun. 2007, 75, 1453–1462. [Google Scholar] [CrossRef]
- Beebe, K.R.; Pell, R.J.; Seasholtz, M.B. Chemometrics: A Practical Guide; Wiley: New York, NY, USA, 1998. [Google Scholar]
- Sharaf, M.A.; Illman, D.L.; Kowalski, B.R. Chemometrics; John Wiley & Sons: New York, NY, USA, 1986. [Google Scholar]
- Westerhuis, J.A.; Kourti, T.; Macgregor, J.F. Analysis of multiblock and hierarchical PCA and PLS models. J. Chemometr. 1998, 12, 301–321. [Google Scholar] [CrossRef]
- Cohen, N.C. Guidebook on Molecular Modeling in Drug Design; Academic Press: San Diego, CA, USA, 1996. [Google Scholar]
- Leach, A.R. Molecular Modeling: Principles and Applications; Prentice Hall: London, UK, 2001. [Google Scholar]
- de Oliveira, T.B. Síntese, elucidação estrutural e avaliação biológica de novos derivados tiazolidínicos e cicloalquil-tiofênicos. M.Sc. Thesis, Graduate Program in Pharmaceutical Sciences. Federal University of Pernambuco, Recife, Brazil, 2010. [Google Scholar]
- Souza, B.C.C. Síntese e Avaliação das atividades Antifúngicas e Antitumorais de novos derivados 2-[(arilideno)amino]-cicloalquil[b]tiofenos-3-carbonitrila. M.Sc. Thesis. Graduate Program in Pharmaceutical Sciences. Federal University of Pernambuco, Recife, Brazil, 2011. [Google Scholar]
- Abu-Hashem, A.A.; El-Shehry, M.F.; Badria, F.A. Design and synthesis of novel thiophenecarbohydrazide, thienopyrazole and thienopyrimidine derivatives as antioxidant and antitumor agents. Acta Pharm. 2010, 60, 311–323. [Google Scholar]
- Cruciani, G.; Crivori, P.; Carrupt, P.-A.; Testa, B. Molecular fields in quantitative structure-permeation relationships: The VolSurf approach. J. Mol. Struct. 2000, 503, 17–30. [Google Scholar]
- Cruciani, G.; Pastor, M.; Guba, W. VolSurf: A new tool for the pharmacokinetic optimization of lead compounds. Eur. J. Pharm. Sci. 2000, 11, S29–S39. [Google Scholar]
- Dragon, Version 5.4; Talete Inc.:: Milano, Italy,, 2006. Available online: http://www.talete.mi.it/products/dragon_description.htm (accessed on 2 January 2012).
- Todeschini, R.; Consonni, V. Handbook of Molecular Descriptors; Wiley-VCH: Weinheim, Germany, 2000. [Google Scholar]
- Golbraikh, A.; Shen, M.; Xiao, Z.; Xiao, Y.-D.; Lee, K.-H.; Tropsha, A. Rational selection of training and test sets for the development of validated QSAR models. J. Comput. Aid. Mol. Des. 2003, 17, 241–253. [Google Scholar] [CrossRef]
- Pastor, M.; Mclay, I.; Pickett, S.; Clementi, S. Grid-Independent descriptors (GRIND): A novel class of alignment-independent three-dimensional molecular descriptors. Med. Chem. 2000, 43, 3233–3243. [Google Scholar] [CrossRef]
- National Committee for Clinical and Laboratory Standards; Method M27-A2; NCCLS: Wayne, MI, USA, 2002; pp. 1–29.
- Nascente, P.S.; Meinerz, A.R.M.; Faria, R.O.; Schuch, L.F.D.; Meireles, M.C.A.; de Mello, J.R.B. CLSI broth microdilution method for testing susceptibility of Malassezia pachydermatis to thiabendazole. Braz. J. Microbiol. 2009, 40, 222–226. [Google Scholar] [CrossRef]
- Barnett, J.A.; Paire, R.W.; Yarrow, D. Yeasts: Characteristics and Identification; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Eloff, J.N. A sensitive and quick microplate method to determine the minimal inhibitory concentration of plant extracts for bacteria. Planta Med. 1978, 64, 711–713. [Google Scholar] [CrossRef]
- Souza, E.L.; Stamford, T.L.M.; Lima, E.O.; Trajano, V.N. Effectiveness of Origanum vulgare L. essential oil to inhibit the growth of food spoiling yeasts. Food Control 2007, 18, 409–413. [Google Scholar] [CrossRef]
- HyperChem, Version 8.0, Hybercube Inc.: Gainesville, FL, USA, 2009.
- Allinger, N.L. A hydrocarbon force-field utilizing V1 and V2 torsional terms. J. Am. Chem. Soc. 1977, 99, 8127–8134. [Google Scholar] [CrossRef]
- Dewar, M.J.S.E.; Zoebisch, G.; Healy, E.F.; Stewart J.J.P. AM1: A new general purpose quantum mechanical molecular model. J. Am. Chem. Soc. 1985, 107, 3902–3909. [Google Scholar] [CrossRef]
- Spartan Model, Version 8.0; Wavefunction, Inc.: Irvine, CA, USA, 2008. Available online: http://www.wavefun.com/products/windows/SpartanModel/win_model.html (accessed on 2 October 2011).
- Zamora, I.; Oprea, T.; Cruciani, G.; Pastor, M.; Ungell, A.-L. Surface descriptors for protein-ligand affinity prediction. J. Med. Chem. 2003, 46, 25–33. [Google Scholar] [CrossRef]
- Crivori, P.; Cruciani, G.; Carrupt, P.-A.; Testa, B. Predicting blood-brain barrier permeation from three-dimensional molecular structure. J. Med. Chem. 2000, 43, 2204–2216. [Google Scholar] [CrossRef]
- Kovatcheva, A.; Golbraikh, A.; Oloff, S.; Xiao, Y.-D.; Zheng, W.; Wolschann, P.; Buchbauer, G.; Tropsha, A. Combinatorial QSAR of ambergris fragrance compounds. J. Chem. Inf. Comp. Sci. 2004, 44, 582–595. [Google Scholar] [CrossRef]
- Oprea, T.I.; Zamora, I.; Ungell, A.-L. Pharmacokinetically based mapping device for chemical space navigation. J. Comb. Chem. 2002, 4, 258–266. [Google Scholar] [CrossRef]
- Cruciani, G.; Pastor, M.; Mannhold, R. Suitability of molecular descriptors for database mining. A comparative analysis. J. Med. Chem. 2002, 45, 2685–2694. [Google Scholar] [CrossRef]
- Cianchetta, G.; Mannhold, R.; Cruciani, G.; Baroni, M.; Cecchetti, V.J. Chemometric studies on the bactericidal activity of quinolones via an extended volsurf approach. J. Med. Chem. 2004, 47, 3193–3201. [Google Scholar] [CrossRef]
- MobyDigs, Version 1.1; Talete Inc.:: Milano, Italy,, 2009. Available online: http://www.talete.mi.it/products/moby_description.htm (accessed on 2 January 2012).
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Scotti, L.; Tullius Scotti, M.; De Oliveira Lima, E.; Sobral da Silva, M.; Do Carmo Alves de Lima, M.; Da Rocha Pitta, I.; Olímpio de Moura, R.; Gonzaga Batista de Oliveira, J.; Duarte da Cruz, R.M.; Bezerra Mendonça, F.J., Júnior. Experimental Methodologies and Evaluations of Computer-Aided Drug Design Methodologies Applied to a Series of 2-Aminothiophene Derivatives with Antifungal Activities. Molecules 2012, 17, 2298-2315. https://doi.org/10.3390/molecules17032298
Scotti L, Tullius Scotti M, De Oliveira Lima E, Sobral da Silva M, Do Carmo Alves de Lima M, Da Rocha Pitta I, Olímpio de Moura R, Gonzaga Batista de Oliveira J, Duarte da Cruz RM, Bezerra Mendonça FJ Júnior. Experimental Methodologies and Evaluations of Computer-Aided Drug Design Methodologies Applied to a Series of 2-Aminothiophene Derivatives with Antifungal Activities. Molecules. 2012; 17(3):2298-2315. https://doi.org/10.3390/molecules17032298
Chicago/Turabian StyleScotti, Luciana, Marcus Tullius Scotti, Edeltrudes De Oliveira Lima, Marcelo Sobral da Silva, Maria Do Carmo Alves de Lima, Ivan Da Rocha Pitta, Ricardo Olímpio de Moura, Jaismary Gonzaga Batista de Oliveira, Rayssa Marques Duarte da Cruz, and Francisco Jaime Bezerra Mendonça, Júnior. 2012. "Experimental Methodologies and Evaluations of Computer-Aided Drug Design Methodologies Applied to a Series of 2-Aminothiophene Derivatives with Antifungal Activities" Molecules 17, no. 3: 2298-2315. https://doi.org/10.3390/molecules17032298