Down Regulation of CIAPIN1 Reverses Multidrug Resistance in Human Breast Cancer Cells by Inhibiting MDR1
Abstract
:1. Introduction
2. Results and Discussion
2.1. Results
2.1.1. Expression and Relationship of P-gp, CIAPIN1 and p53 Protein in Breast Cancer Patient Samples and Cell Lines
2.1.2. Identification of Recombined Plasmid of siRNA Targeting CIAPIN1 and Selection of the Best Interference siRNA Sequence
2.1.3. RNA Interference Test by Lentiviral-Based Vector
2.1.4. CIAPIN1siRNA Reversing the Multidrug Resistance
2.1.5. CIAPIN1siRNA Down Regulate Expression of MDR1
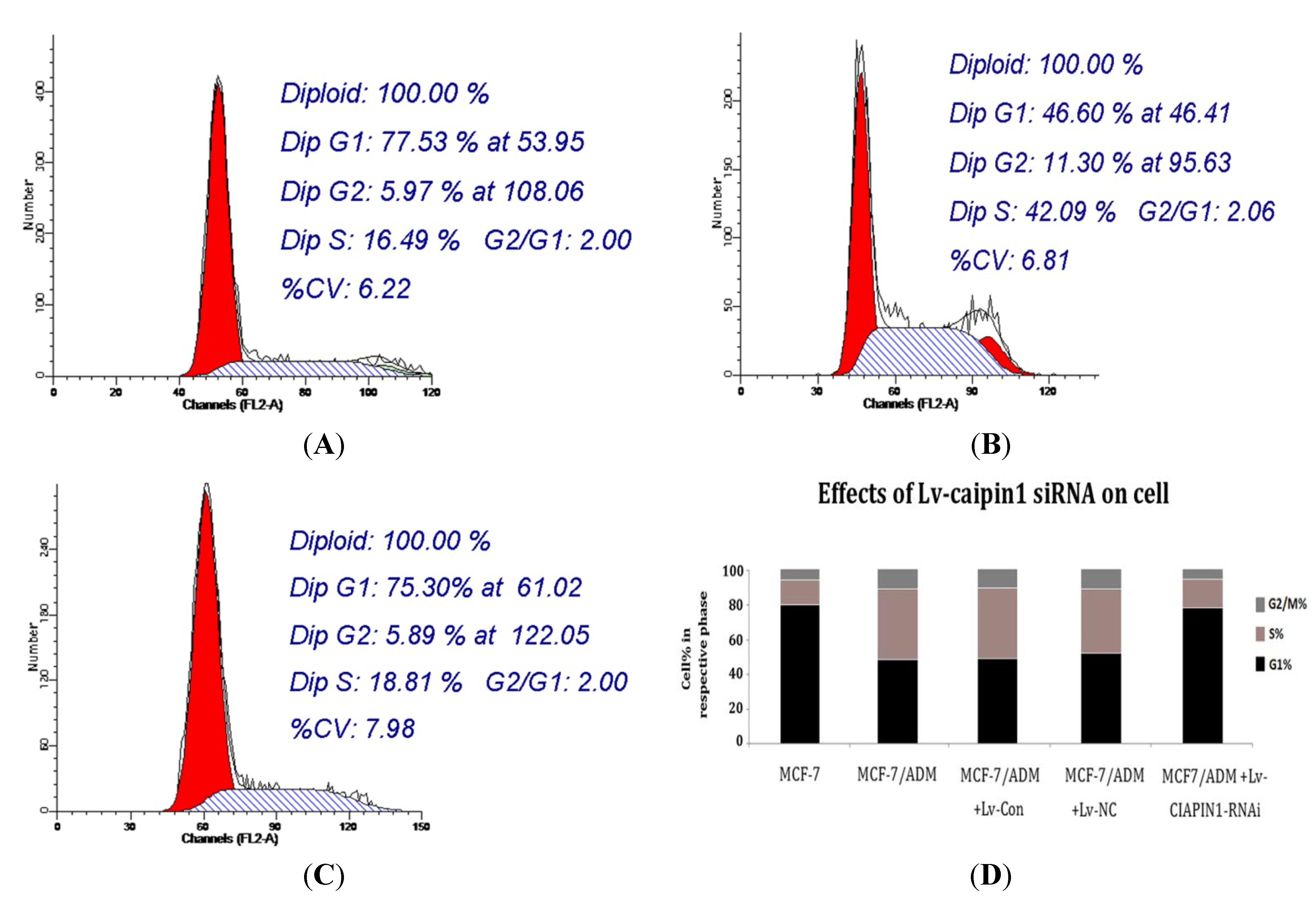
2.1.6. Cell Cycle Analysis
2.1.7. Apoptosis Analysis
2.1.8. CIAPIN1siRNA Influenced the Expression of P53
2.2. Discussion
3. Experimental
3.1. Tissue Samples, Cell Lines and Cell Culture
3.2. Design and Synthesis of Recombined Plasmid of siRNA Targeting CIAPIN1
3.3. Recombinant Lentivirus Generation and Lentivirus Infection
3.4. Real Time-PCR Analysis
3.5. Western Blot Analysis
3.6. Detect IC50 Values of Cells against Various Chemotherapeutics
3.7. Cell Cycle Analysis
3.8. Apoptosis Analysis
3.9. Rhodamine 123 Staining Test
3.10. Statistical Analysis
4. Conclusions
Acknowledgement
References
- Boetes, C. Update on screening breast MRI in high-risk women. Magn. Reson. Imaging Clin. N. Am. 2010, 18, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.B.; Jin, F.; Gao, Y.T. Cancer survival in Shanghai, China, 1992–1995. IARC Sci. Publ. 2011, 162, 55–68. [Google Scholar]
- Seruga, B.; Hertz, P.C.; Le, L.W.; Tannock, I.F. Global drug development in cancer: A cross-sectional study of clinical trial registries. Ann. Oncol. 2010, 21, 895–900. [Google Scholar] [CrossRef] [PubMed]
- Kuo, M.T. Roles of multidrug resistance genes in breast cancer chemoresistance. Adv. Exp. Med. Biol. 2007, 608, 23–30. [Google Scholar] [PubMed]
- Chung, H.C.; Rha, S.Y.; Kim, J.H.; Roh, J.K.; Min, J.S.; Lee, J.S.; Kim, B.S.; Lee, K.B. P-glycoprotein: the intermediate end point of drug response to induction chemo-therapy in locally advanced breast cancer. Breast Cancer Res. Treat. 1997, 42, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Taheri, M.; Mahjoubi, F.; Omranipour, R. Effect of MDR1 polymorphism on multidrug resistance expression in breast cancer patients. Genet. Mol. Res. 2010, 9, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Patwardhan, G.; Gupta, V.; Huang, J.; Gu, X.; Liu, Y.Y. Direct assessment of P-glycoprotein efflux to determine tumor response to chemotherapy. Biochem. Pharmacol. 2010, 80, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Dönmez, Y.; Gündüz, U. Reversal of multidrug resistance by small interfering RNA (siRNA) in doxorubicin-resistant MCF-7 breast cancer cells. Biomed. Pharmacother. 2011, 65, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Shibayama, H.; Takai, E.; Matsumura, I.; Kouno, M.; Morii, E.; Kitamura, Y.; Takeda, J.; Kanakura, Y. Identification of a cytokine-induced antiapoptotic molecule anamorsin essential for definitive hematopoiesis. J. Exp. Med. 2004, 199, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Pan, Y.; Fan, R.; Jin, H.; Han, S.; Liu, J.; Wu, K.; Fan, D. Adenovirus-delivered CIAPIN1 small interfering RNA inhibits HCC growth in vitro and in vivo. Carcinogenesis 2008, 29, 1587–1593. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hao, Z.; Fan, R.; Zou, X.; Jin, H.; Pan, Y.; He, L.; Du, R.; Gao, L.; Liu, D.; et al. CIAPIN1 inhibits gastric cancer cell proliferation and cell cycle progression by downregulating CyclinD1 and upregulating P27. Cancer Biol. Ther. 2007, 6, 1539–1545. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.; Li, X.; Qiao, T.; Li, S.; Lv, Y.; Fan, D. Downregulated expression of CIAPIN1 may contribute to gastric Carciogenesis by accelerating cell proliferation and promoting cell cycle progression. Cancer Biol. 2009, 8, 1064–1070. [Google Scholar] [CrossRef]
- He, J.; Wang, H.; Jin, H.; Guo, C.; Xie, H.; Yan, K.; Li, X.; Shen, Q.; Qiao, T.; Chen, G.; et al. CIAPIN1 inhibits the growth and proliferation of clear cell renal Cell carcinoma. Cancer Lett. 2009, 276, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Shizusawa, T.; Shibayama, H.; Murata, S.; Saitoh, Y.; Sugimoto, Y.; Matsumura, I.; Ogawa, H.; Sugiyama, H.; Fukuhara, S.; Hino, M.; et al. The expression of anamorsin in diffuse large B cell lymphoma: Possible prognostic biomarker for low IPI patients. Leuk. Lymphoma. 2008, 49, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hong, L.; Zhao, Y.; Jin, H.; Fan, R.; Du, R.; Xia, L.; Luo, G.; Fan, D. A new apoptosis inhibitor, CIAPIN1 (cytokine-induced apoptosis inhibitor 1), mediates multidrug resistance in leukemia cells by regulating MDR-1, Bcl-2 and Bax. Biochem. Cell Biol. 2007, 85, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.; Li, X.; Qiao, T.; Du, R.; Hong, L.; Fan, D. CIAPIN1 confers multidrug resistance by upregulating the Expression of MDR-1 and MRP-1 in gastric cancer cells. Cancer Biol. Ther. 2006, 5, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Lage, H. An overview of cancer multidrug resistance: A still unsolved problem. Cell Mol. Life Sci. 2008, 65, 3145–3167. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.K.; Gwak, J.; Song, I.S.; Park, H.S.; Oh, S. Induction of apoptosis in colon cancer cells by a novel topoisomerase I inhibitor TopIn. Biochem. Biophys. Res. Commun. 2011, 409, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Bansal, T.; Akhtar, N.; Jaggi, M.; Khar, R.K.; Talegaonkar, S. Novel formulation approaches for optimising delivery of anticancer drugs based on P-glycoprotein modulation. Drug Discov. Today 2009, 14, 1067–1074. [Google Scholar] [CrossRef] [PubMed]
- Walsh, N.; Larkin, A.; Kennedy, S.; Connolly, L.; Ballot, J.; Ooi, W.; Gullo, G.; Crown, J.; Clynes, M.; O’Driscoll, L. Expression of multidrug resistance markers ABCB1 (MDR-1/P-gp) and ABCC1 (MRP-1) in renal cell carcinoma. BMC Urol. 2009, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Wang, S.; Weng, D.; Chen, G.; Yang, X.; Zhou, J.; Xu, G.; Lu, Y.; Ma, D. Knock-down of P-glycoprotein reverses taxol resistance in ovarian cancer multicellular spheroids. Oncol. Rep. 2007, 17, 117–122. [Google Scholar] [PubMed]
- Hao, Z.; Li, X.; Qiao, T.; Zhang, J.; Shao, X.; Fan, D. Distribution of CIAPIN1 in normal fetal and adult human tissues. J. Histochem. Cytochem. 2006, 54, 417–426. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Wang, H.; Jin, H.; Guo, C.; Xie, H.; Yan, K.; Li, X.; Shen, Q.; Qiao, T.; Chen, G.; et al. CIAPIN1 inhibits the growth and proliferation of clear cell renal Cell carcinoma. Cancer Lett. 2009, 276, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qi, H.W.; Li, X.H.; Chen, X.X.; Liu, J. CIAPIN1 expression in human lung cancer tissues and inhibitory effects of the gene on human pulmonary carcinoma NCI-H446 cells (in Chinese). Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2008, 24, 434–437. [Google Scholar] [PubMed]
- Kuo, M.T. Roles of multidrug resistance genes in breast cancer chemoresistance. Adv. Exp. Med. Biol. 2007, 608, 23–30. [Google Scholar] [PubMed]
- Hannon, G.J. RNA interference. Nature 2002, 418, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Sumimoto, H.; Kawakami, Y. The RNA silencing technology applied by lentiviral vectors in oncology. Methods Mol. Biol. 2010, 614, 187–199. [Google Scholar] [PubMed]
- Sliva, K.; Schnierle, B.S. Selective gene silencing by viral delivery of short hairpin RNA. Virol. J. 2010, 7, 248. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Sun, S.; Oparaocha, I.; Humeau, L.; Davis, B.; Cohen, R.; Binder, G.; Chang, Y.N.; Slepushkin, V.; Dropulic, B. Generation of a packaging cell line for prolonged large- scale production of high-titer HIV-1-based lentiviral vector. J. Gene Med. 2005, 7, 818–834. [Google Scholar] [CrossRef] [PubMed]
- Raoul, C.; Abbas-Terki, T.; Bensadoun, J.C.; Guillot, S.; Haase, G.; Szulc, J.; Henderson, C.E.; Aebischer, P. Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nat. Med. 2005, 11, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Guan, X.; Ma, X. Regulation of IL-27 p28 gene expression in macrophages through MyD88-and interferon-gamma-mediated pathways. J. Exp. Med. 2007, 204, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Gao, H.; Lin, S.Y.; Goss, J.A.; Brunicardi, F.C.; Li, K. siRNA-based targeting of cyclin E overexpression inhibits breast cancer cell growth and suppresses tumor development in breast cancer mouse model. PLoS One 2010, 5, e12860. [Google Scholar] [CrossRef] [PubMed]
- Niu, G.; Castro, C.H.; Nguyen, N.; Sullivan, S.M.; Hughes, J.A. In vitro cytotoxic activity of cationic paclitaxel nanoparticles on MDR-3T3 cells. J. Drug Target. 2010, 18, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Bergman, A.M.; Adema, A.D.; Balzarini, J.; Bruheim, S.; Fichtner, I.; Noordhuis, P.; Fodstad, O.; Myhren, F.; Sandvold, M.L.; Hendriks, H.R.; et al. Antiproliferative activity, mechanism of action and oral antitumor activity of CP-4126, a fatty acid derivative of gemcitabine, in in vitro and in vivo tumor models. Invest. New Drugs 2011, 29, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Mechetner, E.; Kyshtoobayeva, A.; Zonis, S.; Kim, H.; Stroup, R.; Garcia, R.; Parker, R.J.; Fruehauf, J.P. Levels of multidrug resistance (MDR1) P-glycoprotein expression by human breast cancer correlate with in vitro resistance to taxol and doxorubicin. Clin. Cancer Res. 1998, 4, 389–398. [Google Scholar] [PubMed]
- Molnár, J.; Kars, M.D.; Gündüz, U.; Engi, H.; Schumacher, U.; Van Damme, E.J.; Peumans, W.J.; Makovitzky, J.; Gyémánt, N.; Molnár, P. Interaction of tomato lectin with ABC transporter in cancer cells: Glycosylation confers functional conformation of P-gp. Acta Histochem. 2009, 111, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, H.; Reagan-Shaw, S.; Breur, J.; Ahmad, N. Sanguinarine induces apoptosis of human pancreatic carcinoma AsPC-1 and BxPC-3 cells via modulations in Bcl-2 family proteins. Cancer Lett. 2007, 249, 198–208. [Google Scholar] [CrossRef] [PubMed]
- Holcomb, B.; Yip-Schneider, M.; Schmidt, C.M. The role of nuclear factor kappaB in pancreatic cancer and the clinical applications of targeted therapy. Pancreas 2008, 36, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Xie, Z.H.; Cai, G.P.; Jiang, Y.Y. The effect of survivin on multidrug resistance mediated by P-glycoprotein in MCF-7 and its adriamycin resistant cells. Biol. Pharm. Bull. 2007, 30, 2279–2283. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, D.; Rotter, V. p53-dependent cell cycle control: Response to genotoxic stress. Semin. Cancer Biol. 1998, 8, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Oue, T.; Yoneda, A.; Uehara, S.; Yamanaka, H.; Fukuzawa, M. Increased expression of multidrug resistance-associated genes after chemotherapy in pediatric solid malignancies. J. Pediatr. Surg. 2009, 44, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.F.; Li, X.H.; Shi, Y.Q.; Wu, Y.Y.; Li, N.; He, Q.; Ji, Q.; Wang, R.Q.; Yang, S.M.; Fang, D.C. CIAPIN1 confers multidrug resistance through up-regulation of MDR-1 and Bcl-L in LoVo/Adr cells and is independent of p53. Oncol. Rep. 2011, 25, 1091–1098. [Google Scholar] [PubMed]
Sample Availability: Not available. |









| P-gp | CIAPIN1 | p53 | ||||
|---|---|---|---|---|---|---|
| + | − | P | + | − | P | |
| + | 11 | 10 | <0.01 | 13 | 8 | <0.025 |
| − | 3 | 17 | 4 | 16 | ||
| Group | IC50 value | ||
|---|---|---|---|
| paclitaxel | epirubicin | gemcitabine | |
| MCF7 | 0.808 ± 0.004 | 1.067 ± 0.016 | 5.859 ± 0.551 |
| MCF7/ADM | 7.121 ± 0.312 | 11.206 ± 1.789 | 49.724 ± 4.522 |
| MCF7/ADM + CIAPIN1-RNAi | 1.134 ± 0.057 * | 4.514 ± 0.203 * | 18.298 ± 1.273 * |
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lu, D.; Xiao, Z.; Wang, W.; Xu, Y.; Gao, S.; Deng, L.; He, W.; Yang, Y.; Guo, X.; Wang, X. Down Regulation of CIAPIN1 Reverses Multidrug Resistance in Human Breast Cancer Cells by Inhibiting MDR1. Molecules 2012, 17, 7595-7611. https://doi.org/10.3390/molecules17067595
Lu D, Xiao Z, Wang W, Xu Y, Gao S, Deng L, He W, Yang Y, Guo X, Wang X. Down Regulation of CIAPIN1 Reverses Multidrug Resistance in Human Breast Cancer Cells by Inhibiting MDR1. Molecules. 2012; 17(6):7595-7611. https://doi.org/10.3390/molecules17067595
Chicago/Turabian StyleLu, Dan, Zhibo Xiao, Wenxiu Wang, Yuqing Xu, Shujian Gao, Lili Deng, Wen He, Yu Yang, Xiaofei Guo, and Xuemei Wang. 2012. "Down Regulation of CIAPIN1 Reverses Multidrug Resistance in Human Breast Cancer Cells by Inhibiting MDR1" Molecules 17, no. 6: 7595-7611. https://doi.org/10.3390/molecules17067595
APA StyleLu, D., Xiao, Z., Wang, W., Xu, Y., Gao, S., Deng, L., He, W., Yang, Y., Guo, X., & Wang, X. (2012). Down Regulation of CIAPIN1 Reverses Multidrug Resistance in Human Breast Cancer Cells by Inhibiting MDR1. Molecules, 17(6), 7595-7611. https://doi.org/10.3390/molecules17067595




