Investigation of Non-Enzymatic Glycosylation of Human Serum Albumin Using Ion Trap-Time of Flight Mass Spectrometry
Abstract
:1. Introduction
2. Results and Discussion
2.1. GA Purification and GA Level in Serum
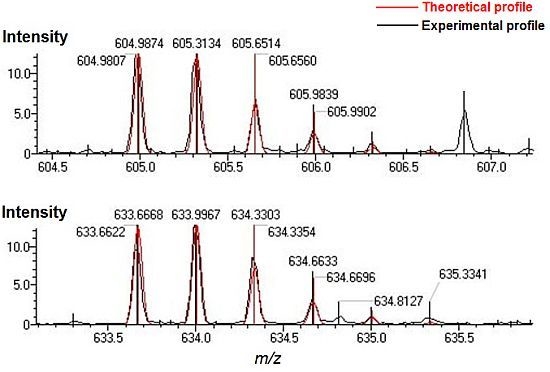
2.2. Glycation Site Determination
2.3. Incorporation Ratio of Glucose to Albumin
3. Experimental
3.1. Materials
3.2. Sample Preparation
3.3. Affinity Chromatography
3.4. SDS-PAGE Analysis
3.5. Enzymatic Digestion
3.6. LC/MS/MS Analysis
3.7. Data Processing
3.8. MALDI-TOF-MS Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
References
- Rondeau, P.; Bourdon, E. The glycation of albumin: Structural and functional impacts. Biochimie 2011, 93, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Day, J.F.; Thorpe, S.R.; Baynes, J.W. Nonenzymatically glucosylated albumin. In vitro preparation and isolation from normal human serum. J. Biol. Chem. 1979, 254, 595–597. [Google Scholar] [PubMed]
- Sattarahmady, N.; Moosavi-Movahedi, A.A.; Ahmad, F.; Hakimelahi, G.H.; Habibi-Rezaei, M.; Saboury, A.A.; Sheibani, N. Formation of the molten globute-like state during prolonged glycation of human serum albumin. Biochim. Biophys. Acta 2007, 1770, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Shaklai, N.; Garlick, R.L.; Bunn, H.F. Nonenzymatic glycosylation of human serum albumin alters its conformation and function. J. Biol. Chem. 1984, 259, 3812–3817. [Google Scholar] [PubMed]
- Takino, J.; Yamagishi, S.; Takeuchi, M. Cancer malignancy is enhanced by glyceraldehyde-derived advanced glycation end-products. J. Oncol. 2010, 2010, 739852. [Google Scholar] [CrossRef] [PubMed]
- Peppa, M.; Raptis, S.A. Advanced glycation end products and cardiovascular disease. Curr. Diabetes Rev. 2008, 4, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Münch, G.; Deuther-Conrad, W.; Gasic-Milenkovic, J.J. Glycoxidative stress creates a vicious cycle of neurodegeneration in Alzheimer’s disease-a target for neuroprotective treatment strategies? Neural. Transm. Suppl. 2002, 62, 303–307. [Google Scholar]
- Roohk, H.V.; Zaidi, A.R. A review of glycated albumin as an intermediate glycation index for controlling diabetes. J. Diabetes Sci. Technol. 2008, 2, 1114–1121. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Uchino, H.; Shimizu, T.; Kanazawa, A.; Tamura, Y.; Sakai, K.; Watada, H.; Hirose, T.; Kawamori, R.; Tanaka, Y. Comparison of glycated albumin (GA) and glycated hemoglobin (HbAlc) in type 2 diabetic patients: Usefulness of GA for evaluation of short term changes in glycemic control. Endocr. J. 2007, 54, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Yatscoff, R.W.; Tevaarwerk, G.J.M.; MacDonald, J.C. Quantification of nonenzymically glycated albumin and total serum protein by affinity chromatography. Clin. Chem. 1984, 30, 446–449. [Google Scholar] [PubMed]
- Zhang, G.; Kai, M.; Nohta, H.; Umegae, Y.; Ohkura, Y. Simultaneous determination of glycated albumin and D-Glucose in human serum by high-performance liquid chromatography with postcolumn fluorescence derivatization. Anal. Sci. 1993, 9, 9–14. [Google Scholar] [CrossRef]
- Lapolla, A.; Fedele, D.; Reitano, R.; Aricò, N.C.; Seraglia, R.; Traldi, P.; Marotta, E.; Tonani, R. Enzymatic digestion and mass spectrometry in the study of advanced glycation end products/peptides. J. Am. Soc. Mass Spectrom. 2004, 15, 496–509. [Google Scholar] [CrossRef] [PubMed]
- Mashiba, S.; Uchida, K.; Okuda, S.; Tomita, S. Measurement of glycated albumin by the nitroblue tetrazolium colorimetric method. Clin. Chim. Acta 1992, 212, 3–15. [Google Scholar] [CrossRef]
- Šenfeld, A.; Pavlíček, Z. Nonenzymatic glycosylation of human serum albumin: Fluorescence and chemiluminescence behavior. J. Fluoresc. 1993, 3, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Yasukawa, K.; Abe, F.; Shida, N.; Koizumi, Y.; Uchida, T.; Noguchi, K.; Shima, K. High-performance affinity chromatograph system for the rapid, efficient assay of glycated albumin. J. Chromatogr. 1992, 597, 271–275. [Google Scholar] [CrossRef]
- Iberg, N.; Flückiger, R. Nonenzymatic glycosylation of albumin in vivo. Identification of multiple glycosylated sites. J. Biol. Chem. 1986, 261, 13542–13545. [Google Scholar] [PubMed]
- Garlick, R.L.; Mazer, J.S. The principal site of nonenzymatic glycosylation of human serum albumin in vivo. J. Biol. Chem. 1983, 258, 6142–6146. [Google Scholar] [PubMed]
- Zmatliková, Z.; Sedláková, P.; Lacinová, K.; Eckhardt, A.; Pataridis, S.; Mikšík, I. Non-enzymatic posttranslational modifications of bovine serum albumin by oxo-compounds investigated by high-performance liquid chromatography-mass spectrometry and capillary zone electrophoresis-mass spectrometry. J. Chromatogr. A 2010, 1217, 8009–8015. [Google Scholar] [CrossRef] [PubMed]
- Zoellner, H.; Hou, J.Y.; Hochgrebe, T.; Poljak, A.; Duncan, M.W.; Golding, J.; Henderson, T.; Lynch, G. Fluorometric and mass spectrometric analysis of nonenzymatic glycosylated albumin. Biochem. Biophys. Res. Commun. 2001, 284, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Kisugi, R.; Kouzuma, T.; Yamamoto, T.; Akizuki, S.; Miyamoto, H.; Someya, Y.; Yokoyama, J.; Abe, I.; Hirai, N.; Ohnishi, A. Structural and glycation site changes of albumin in diabetic patient with very high glycated albumin. Clin. Chim. Acta 2007, 382, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Frolov, A.; Hoffmann, R. Identification and relative quantification of specific glycation sites in the human serum albumin. Anal. Bioanal. Chem. 2010, 397, 2349–2356. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.M.; Collings, B.A.; Douglas, D.J. A new linear ion trap time-of-flight system with tandem mass spectrometry capabilities. Rapid Commun. Mass Spectrom. 1998, 12, 1463–1474. [Google Scholar] [CrossRef]
- Collings, B.A.; Campbell, J.M.; Mao, D.; Douglas, D.J. A combined linear ion trap time-of-flight system with improved performance and MSn capabilities. Rapid Commun. Mass Spectrom. 2001, 15, 1777–1795. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Jin, X.; Misek, D.; Hinderer, R.; Hanash, S.M.; Lubman, D.M. Identification of proteins from two-dimensional gel electrophoresis of human erythroleukemia cells using capillary high performance liquid chromatography/electrospray-ion trap-reflection time-of-flight mass spectrometry with two-dimensional topographic map analysis of in-gel tryptic digest products. Rapid Commun. Mass Spectrom. 1999, 13, 1907–1916. [Google Scholar] [PubMed]
- Huang, P.; Wall, D.B.; Parus, S.; Lubman, D.M. On-line capillary liquid chromatography tandem mass spectrometry on an Ion trap/reflection time-of-flight mass spectrometer using the sequence tag database search approach for peptide sequencing and protein identification. J. Am. Soc. Mass Spectrom. 2000, 11, 127–135. [Google Scholar] [CrossRef]
- Hyakkoku, K.; Hamanaka, J.; Tsuruma, K.; Shimazawa, M.; Hara, H. Proteomic approach with LCMS-IT-TOF identified an increase of Rab33B after transient focal cerebral ischemia in mice. Exp. Transl. Stroke Med. 2010, 2, 20. [Google Scholar] [CrossRef] [PubMed]
- Skinner, J.P.; Chi, L.; Ozeata, P.F.; Ramsay, C.S.; ÓHara, R.L.; Calfin, B.B.; Tetin, S.Y. Introduction of the mass spread function for characterization of protein conjugates. Anal. Chem. 2012, 84, 1172–1177. [Google Scholar] [CrossRef] [PubMed]
- Kratz, F. Albumin as a drug carrier: Design of prodrugs, drug conjugates and nanoparticles. J. Control. Release 2008, 132, 171–183. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the GA and NGA are available from the authors. |





| Position | Peptide sequence | m/z calculated | m/z measured | error (ppm) |
|---|---|---|---|---|
| T2-3 | SEVAHRFK | 487.2643 | 487.2728 | 17.38 |
| T3-4 | FKDLGEENFK | 409.5399 | 409.5366 | −8.08 |
| T4 | DLGEENFK | 476.2245 | 476.2165 | −16.88 |
| T5 | ALVLIAFAQYLQQCPFEDHVK | 830.7665 | 830.7538 | −15.3 |
| T6 | LVNEVTEFAK | 575.3111 | 575.3013 | −17.11 |
| T8 | SLHTLFGDK | 509.2718 | 509.2650 | −13.4 |
| T9 | LCTVATLR | 467.2629 | 467.2547 | −17.65 |
| T10 | ETYGEMADCCAK | 717.7703 | 717.7604 | −13.86 |
| gT10-11 | ETYGEMADCCAKQEPER | 745.9661 | 745.9593 | −9.17 |
| T12-13 | NECFLQHKDDNPNLPR | 666.3146 | 666.3077 | −10.42 |
| T14 | LVRPEVDVMCTAFHDNEETFLK | 884.0928 | 884.0804 | −14.08 |
| T15-16 | KYLYEIAR | 528.2978 | 528.2894 | −15.98 |
| T16 | YLYEIAR | 464.2504 | 464.2443 | −13.05 |
| T17-18 | RHPYFYAPELLFFAK | 633.6699 | 633.6621 | −12.34 |
| T18 | HPYFYAPELLFFAK | 581.6362 | 581.6272 | −15.5 |
| T18-19 | HPYFYAPELLFFAKR | 633.6699 | 633.6621 | −12.34 |
| gT24-26 | DEGKASSAKQR | 669.8284 | 669.8179 | −15.71 |
| gT28-29 | CASLQKFGER | 679.3245 | 679.3377 | 19.49 |
| T29 | FGER | 508.2514 | 508.2404 | −21.69 |
| T30-31 | AFKAWAVAR | 510.2929 | 510.2822 | −20.96 |
| gT32-34 | LSQRFPKAEFAEVSK | 633.6668 | 633.6622 | −7.25 |
| T33-34 | FPKAEFAEVSK | 626.8322 | 626.8340 | 2.8 |
| gT33-34 | FPKAEFAEVSK | 472.2415 | 472.2353 | −13.19 |
| T34 | AEFAEVSK | 440.7242 | 440.7256 | 3.23 |
| T34-35 | AEFAEVSKLVTDLTK | 550.9698 | 550.9661 | −6.74 |
| gT34-35 | AEFAEVSKLVTDLTK | 604.9874 | 604.9807 | −11.11 |
| T36 | VHTECCHGDLLECADDR | 696.2840 | 696.2741 | −14.26 |
| gT37-38 | ADLAKYICENQDSISSK | 701.9965 | 701.9827 | −19.71 |
| T38 | YICENQDSISSK | 722.3247 | 722.3142 | −14.48 |
| T39-40 | LKECCEKPLLEK | 516.2704 | 516.2639 | −12.66 |
| T40 | ECCEKPLLEK | 653.3125 | 653.3032 | −14.24 |
| T41 | SHCIAEVENDEMPADLPSLA ADFVESK | 992.1197 | 992.1038 | −16.0 |
| T44 | DVFLGMFLYEYAR | 812.3974 | 812.3844 | −16.04 |
| T45-46 | RHPDYSVVLLLR | 489.9525 | 489.9460 | −13.34 |
| T46 | HPDYSVVLLLR | 437.9188 | 437.9120 | −15.6 |
| gT47-48 | LAKTYETTLEK | 486.9240 | 486.9212 | −5.71 |
| T50 | VFDEFKPLVEEPQNLIK | 682.3700 | 682.3583 | −17.11 |
| gT50 | VFDEFKPLVEEPQNLIK | 736.3876 | 736.3848 | −3.78 |
| T51 | QNCELFEQLGEYK | 829.3800 | 829.3642 | −19.01 |
| gT53-54 | YTKK | 701.3716 | 701.3727 | 1.57 |
| T54-T55 | KVPQVSTPTLVEVSR | 547.3174 | 547.3109 | −11.94 |
| T55 | VPQVSTPTLVEVSR | 756.4250 | 756.4124 | −16.7 |
| gT56-57 | NLGKVGSK | 482.7691 | 482.7736 | 9.29 |
| T57-59 | VGSKCCKHPEAK | 700.8423 | 700.8282 | −20.11 |
| T61 | MPCAEDYLSVVLNQLCVLHEK | 840.0761 | 840.0623 | −16.47 |
| gT62-64 | TPVSDRVTKCCTESLVNR | 762.0351 | 762.0489 | 18.1 |
| T64 | CCTESLVNR | 569.7526 | 569.7459 | −11.79 |
| T65 | RPCFSALEVDETYVPK | 637.6487 | 637.6394 | −14.65 |
| T66 | EFNAETFTFHADICTLSEK | 754.0124 | 753.9980 | −19.11 |
| gT68-70 | QIKKQTALVELVK | 554.0012 | 553.9944 | −12.31 |
| T69-70 | KQTALVELVK | 564.8530 | 564.8464 | −11.65 |
| gT69-70 | KQTALVELVK | 645.8794 | 645.8683 | −17.18 |
| T70 | QTALVELVK | 500.8055 | 500.7996 | −11.78 |
| gT73-74 | EQLKAVMDDFAAFVEK | 668.3274 | 668.3174 | −15.03 |
| T74 | AVMDDFAAFVEK | 671.8210 | 671.8094 | −17.3 |
| gT74-75 | AVMDDFAAFVEKCCK | 651.6195 | 651.6146 | −7.47 |
| gT76-77 | ADDKETCFAEEGK | 554.5664 | 554.5763 | 17.83 |
| T78-79 | KLVAASQAALGL | 571.3506 | 571.3419 | −15.24 |
| T79 | LVAASQAALGL | 507.3031 | 507.2958 | −14.44 |
| Subject | Glycation sites |
|---|---|
| healthy person | Lys-64, Lys-93*, Lys-190 or Lys-195*, Lys-205*, Lys-225*, Lys-233, Lys-262, Lys-274, Lys-281, Lys-323*, Lys-351, Lys-378, Lys-413*, Lys-432*, Lys-475, Lys-525, Lys-526*, Lys-545, Lys-557*, Lys-564*, Lys-573 or Lys-574* |
| diabetic patient | Lys-64, Lys-190*, Lys-199, Lys-225*, Lys-233, Lys-240*, Lys-274, Lys-281, Lys-281 or Lys-286, Lys-286*, Lys-317, Lys-323*, Lys-372*, Lys-413*, Lys-475, Lys-525, Lys-557 or Lys-560 or Lys-564* |
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Bai, X.; Wang, Z.; Huang, C.; Wang, Z.; Chi, L. Investigation of Non-Enzymatic Glycosylation of Human Serum Albumin Using Ion Trap-Time of Flight Mass Spectrometry. Molecules 2012, 17, 8782-8794. https://doi.org/10.3390/molecules17088782
Bai X, Wang Z, Huang C, Wang Z, Chi L. Investigation of Non-Enzymatic Glycosylation of Human Serum Albumin Using Ion Trap-Time of Flight Mass Spectrometry. Molecules. 2012; 17(8):8782-8794. https://doi.org/10.3390/molecules17088782
Chicago/Turabian StyleBai, Xue, Zhangjie Wang, Chengcai Huang, Zhe Wang, and Lianli Chi. 2012. "Investigation of Non-Enzymatic Glycosylation of Human Serum Albumin Using Ion Trap-Time of Flight Mass Spectrometry" Molecules 17, no. 8: 8782-8794. https://doi.org/10.3390/molecules17088782