Synthesis and Characterization of Cationic Glycidyl-Based Poly(aminoester)-Folic Acid Targeting Conjugates and Study on Gene Delivery
Abstract
:1. Introduction
2. Results and Discussion
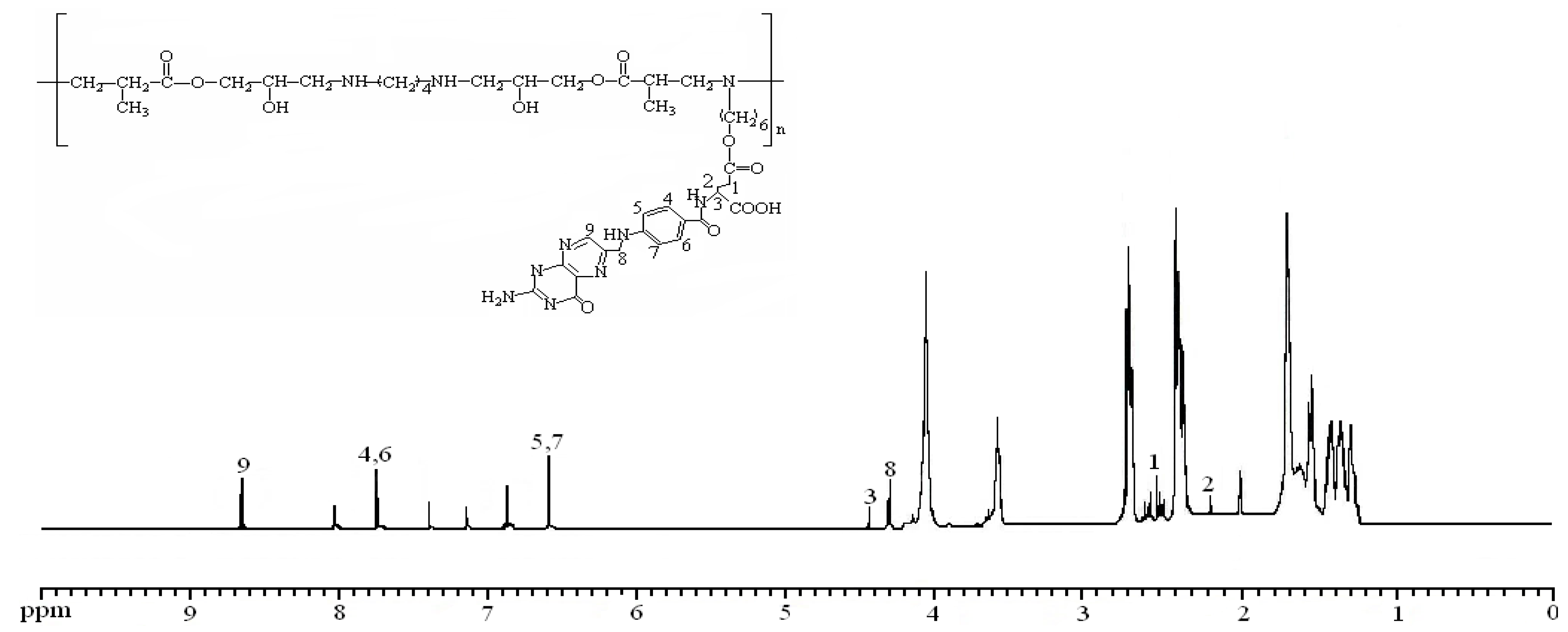
2.1. Structural Characterizations of EPAE-FA

2.2. Size and Zeta-Potential Analysis of Polymer/DNA Complexes


2.3. Buffering Capacity of Polymers

2.4. Cytotoxicity of Polymers/DNA


2.5. DNA Gel Retardation Assay

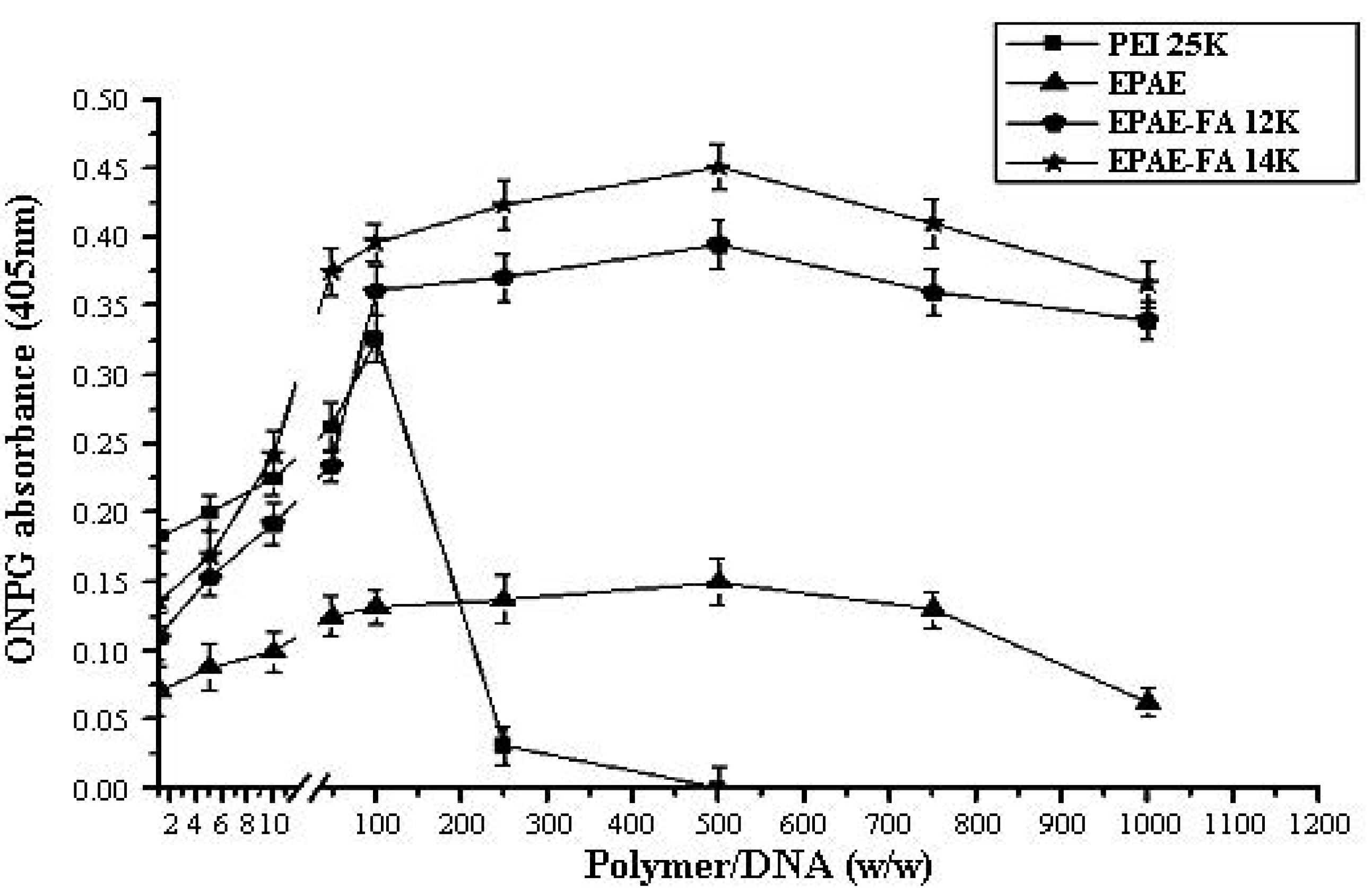
2.6. Cellular Delivery of Plasmid DNA via EPAE-FA Vectors



3. Experimental
3.1. Materials
3.2. Polymer Characterizations
3.3. Synthesis of EPAE-FA12k (EPAE-FA14k)
3.4. Characterization of EPAE Moiety of EPAE-FA
3.5. Characterization of FA Moiety of EPAE-FA
 ), 2.15 (2H, –OC(O)CH2CH2CH– (COOH)–), 4.29 (2H, –OC(O)CH2CH2CH(COOH)–), 4.52(1H, –OC(O)CH2CH2CH(COOH)–), 6.65 (2H,
), 2.15 (2H, –OC(O)CH2CH2CH– (COOH)–), 4.29 (2H, –OC(O)CH2CH2CH(COOH)–), 4.52(1H, –OC(O)CH2CH2CH(COOH)–), 6.65 (2H,  ), 7.71 (2H,
), 7.71 (2H,  ), 8.05 (1H, –CH2CH2CH(COOH)NH-), 8.65(1H,
), 8.05 (1H, –CH2CH2CH(COOH)NH-), 8.65(1H,  ).
). 3.6. Acid-Base Titration Assay of Polymers
3.7. XTT Assay
3.8. Formation of Polymer/DNA Complexes
3.9. Characterizations of Polymer/DNA Complexes
3.10. DNA Gel Retardation Assay of Polymer/DNA Complexes
3.11. Transfection Protocol and ONPG Assay
4. Conclusions
Acknowledgments
Conflict of Interest
References
- EI-Aneed, A. An overview of current delivery systems in cancer gene therapy. J. Control. Release 2004, 94, 1–14. [Google Scholar] [CrossRef]
- Olefsky, J.M. Diabetes gene therapy for rats and mice. Nature 2000, 408, 420–421. [Google Scholar] [CrossRef]
- Thomas, C.E.; Ehrhardt, A.; Kay, M.A. Progress and problem with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003, 4, 346–358. [Google Scholar] [CrossRef]
- Pfeifer, A.; Verma, I.M. Gene therapy: Promises and problems. Annu. Rev. Genomics Hum. Genet. 2001, 2, 177–211. [Google Scholar] [CrossRef]
- Behr, J.P. The proton sponge: A trick to enter cells the viruses did not exploit. Chimia 1997, 51, 34–36. [Google Scholar]
- Boussif, O.; Lezoualc’h, F.; Zanta, M.A.; Mergny, M.D.; Scherman, D.; Demeneix, B.; Behr, J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in-vivo-polyethylenimine. Proc. Natl. Acad. Sci. USA 1995, 92, 7297–7301. [Google Scholar]
- Asayama, S.; Maruyama, A.; Cho, C.S.; Akaike, T. Design of comb-type polyamine copolymers for a novel pH-sensitive DNA carrier. Bioconjug. Chem. 1997, 8, 833–838. [Google Scholar] [CrossRef]
- Van de Wetering, P.; Cherng, J.Y.; Talsma, H.; Hennink, W.E. Relation between transfection efficiency and cytotoxicity of poly(2-(dimethylamino)ethyl methacrylate)/plasmid nanoparticles. J. Control. Release 1997, 49, 59–69. [Google Scholar] [CrossRef]
- Zhang, X.Q.; Wang, X.L.; Huang, S.W.; Zhuo, R.X.; Liu, Z.L.; Mao, H.Q.; Leong, K.W. In vitro gene delivery using polyamidoamine dentrimers with a trimesyl core. Biomacromolecules 2005, 6, 341–350. [Google Scholar] [CrossRef]
- Putnam, D.; Langer, R. Poly(4-hydroxy-L-proline ester): Low-temperature polycondensation and plasmid DNA complexation. Macromolecules 1999, 32, 3658–3662. [Google Scholar] [CrossRef]
- Lim, Y.B.; Kim, C.H.; Kim, K.; Kim, S.W.; Park, J.S. Development of a safe gene delivery system using biodegradable polymer, poly(alpha-(4-aminobutyl)-L-glycolic acid). J. Am. Chem. Soc. 2000, 122, 6524–6525. [Google Scholar] [CrossRef]
- Lim, Y.B.; Kim, S.M.; Suh, H.; Park, J.S. Biodegradable, endosome disruptive, and cationic network-type polymers as a highly efficient and nontoxic gene delivery carrier. Bioconjug. Chem. 2002, 13, 952–957. [Google Scholar] [CrossRef]
- Lin, C.H.; Hsiao, Y.C.; Shau, M.D. Synthesis and characterizations of new glycidyl-based cationic poly(aminoester) and study on gene delivery. Int. J. Pharm. 2010, 393, 136–142. [Google Scholar] [CrossRef]
- Guo, K.F.; Shi, S.; Wang, X.H.; Kan, B.; Gu, Y.C.; Shi, X.Y.; Luo, F.; Zhao, X.; Wei, Y.Q.; Qian, Z.Y. Synthesis of a novel biodegradable poly(esteramine)(PEAs) copolymer based on low-molecular-weight polyethylenimine for gene delivery. Int. J. Pharm. 2009, 379, 82–89. [Google Scholar] [CrossRef]
- Akinc, A.; Anderson, D.G.; Lynn, D.M.; Langer, R. Synthesis of poly(beta-amino ester)s optimized for high effective gene delivery. Bioconjug. Chem. 2003, 14, 979–988. [Google Scholar] [CrossRef]
- Lynn, D.M.; Anderson, D.G.; Putnam, D.; Langer, R. Accelerated discovery of synthetic transfection vectors: Parallel synthesis and screening of degradable polymer library. J. Am. Chem. Soc. 2001, 123, 8155–8156. [Google Scholar]
- Wang, J.; Huang, S.W.; Zhang, P.C.; Mao, H.Q.; Leong, K.W. Effect of side-chain structure on gene transfer efficiency of biodegradable cationic polyphosphoesters. Int. J. Pharm. 2003, 265, 75–84. [Google Scholar] [CrossRef]
- Zhao, Z.; Wang, J.; Mao , H.Q.; Leong, K.W. Polyphosphoesters in drug and gene delivery. Adv.Drug Deliv.Rev. 2003, 55, 483–499. [Google Scholar] [CrossRef]
- Liu, X.Y.; Ho, W.Y.; Hung, W.J.; Shau, M.D. The characteristics and transfection efficiency of cationic poly(ester-co-urethane)-short chain pei conjugates self-assembled with DNA. Biomaterials 2009, 30, 6665–6673. [Google Scholar] [CrossRef]
- Jian, Z.Y.; Chang, J.K.; Shau, M.D. Synthesis and characterizations of new lysine-based biodegradable cationic poly(urethane-co-ester) and study on self-assembled nano-particles with DNA. Bioconjug. Chem. 2009, 20, 774–779. [Google Scholar] [CrossRef]
- Ahn, C.H.; Chae, S.Y.; Bae, Y.H.; Kim, S.W. Biodegradable poly(ethylenimine) for plasmid delivery. J. Control. Release 2002, 80, 273–282. [Google Scholar] [CrossRef]
- Petersen, H.; Merdan, T.; Kunath, F.; Fischer, D.; Kissel, T. Poly(ethylenimine-co-l-lactamide-co-succinamide): A biodegradable polyethylenimine derivative with an advantageous pH-dependent hydrolytic degradation for gene delivery. Bioconjug. Chem. 2002, 13, 812–821. [Google Scholar] [CrossRef]
- Lu, Y.; Low, P.S. Floate-mediated delivery of macromolecular anticancer therapeutic agents. Adv. Drug Deliv. Rev. 2002, 54, 675–693. [Google Scholar] [CrossRef]
- Cheng, H.; Zhu, J.L.; Zeng, X.; Jing, Y.; Zhang, X.Z.; Zhuo, R.X. Targeted gene delivery mediated by floate-polyethylenimine-block-poly(ethylene glycol) with receptor selectivity. Bioconjug. Chem. 2009, 20, 481–487. [Google Scholar] [CrossRef]
- Liang, B.; He, M.L.; Xiao, Z.P.; Li, Y.; Chan, C.; Kung, H.F.; Shuai, X.T.; Peng, Y. Synthesis and characterization of floate-PEG-grafted-hyperbranched-PEI for tumor-targeted gene delivery. Biochem. Biophys. Res. Commun. 2008, 367, 874–880. [Google Scholar] [CrossRef]
- Liu, S.Q.; Wiradharma, N.; Gao, S.J.; Tong, Y.W.; Yang, Y.Y. Bio-functional micelles self-assembled from a floate-conjugated block copolymer for targeted intracellular delivery of anticancer drugs. Biomaterials 2007, 28, 1423–1433. [Google Scholar] [CrossRef]
- Sample Availability: Not available.
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Tsai, P.F.; Chang, W.Y.; Hsiao, Y.C.; Li, K.J.; Shau, M.D. Synthesis and Characterization of Cationic Glycidyl-Based Poly(aminoester)-Folic Acid Targeting Conjugates and Study on Gene Delivery. Molecules 2012, 17, 9056-9069. https://doi.org/10.3390/molecules17089056
Tsai PF, Chang WY, Hsiao YC, Li KJ, Shau MD. Synthesis and Characterization of Cationic Glycidyl-Based Poly(aminoester)-Folic Acid Targeting Conjugates and Study on Gene Delivery. Molecules. 2012; 17(8):9056-9069. https://doi.org/10.3390/molecules17089056
Chicago/Turabian StyleTsai, Pai Feng, Wei Yang Chang, Yu Che Hsiao, Kuo Jung Li, and Min Da Shau. 2012. "Synthesis and Characterization of Cationic Glycidyl-Based Poly(aminoester)-Folic Acid Targeting Conjugates and Study on Gene Delivery" Molecules 17, no. 8: 9056-9069. https://doi.org/10.3390/molecules17089056



