Microfluidic Approaches to Bacterial Biofilm Formation
Abstract
:1. Introduction
2. Biofilm Development
2.1. Why Do Planktonic Cells form Biofilms?
2.2. Biofilm Development Sequence

2.3. Determinants of Biofilm Development
2.3.1. Gene Expression
2.3.2. Surface Properties
2.3.3. Hydrodynamic Conditions
2.3.4. Quorum Sensing Signals
2.3.5. Characteristics of the Aqueous Medium
| Environmental conditions | Effect on biofilms | Species | Reference |
|---|---|---|---|
| Surface properties | |||
| surface roughness | Positive | P. aeruginosa | [47] |
| hydrophobicity | Positive | Pseudomonas sp. | [47] |
| non-polar surface | Positive | Pseudomonas sp. | [48] |
| porosity | Positive | Corynebacterium, Rhodococcus, Gordona | [49,50] |
| cations on the surface | Positive | P. fluorescens | [51] |
| chloropropyl-terminated surface | Positive | P. fluorescens | [52] |
| alkyl-terminated surface | Negative | P. fluorescens | [52] |
| nanostructure of the surface | Positive/Negative | S. aureus, S. epidermidis, P. aeruginosa | [10] |
| Hydrodynamic conditions | |||
| residence time | Positive | P. aeruginosa | [24] |
| shear stress at the interface | Positive/Negative | P. aeruginosa, P. fluorescens | [54,55,56] |
| hetero-stress distribution at the interface | Negative | P. aeruginosa | [25] |
| Quorum sensing signals | |||
| quorum sensing autoinducers | Positive | Gram-negative bacteria | [57] |
| Characteristics of the aqueous medium | |||
| nutrient source | Positive/Negative | P. aeruginosa | [60] |
| nutrient starvation | Negative | P. aeruginosa | [61] |
| oxygen concentration in the fluid | Positive | P. aeruginosa | [13] |
| carbon dioxide concentration in the fluid | Positive | P. putida | [62] |
| dense phase carbon dioxide | Negative | P. aeruginosa | [63] |
3. Microfluidic Approach
3.1. Advantages of Microfluidics

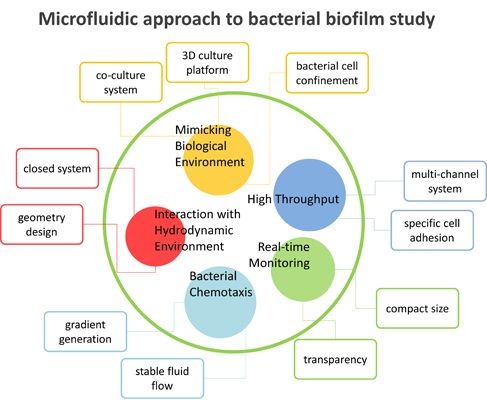
3.2. Microfluidics Approaches in Bacterial Biofilm Studies
3.2.1. Interaction with Hydrodynamic Environment
3.2.2. Bacterial Chemotaxis
3.2.3. High-Throughput Analysis

3.2.4. Real-Time Monitoring
| Analysis techniques | Microfluidic approach | Acquired information | Reference |
|---|---|---|---|
| Fluorescence microscopy | generating chemical (antibiotic) gradient | antibiotic susceptibility of bacterial biofilms | [79] |
| Confocal reflection microscopy | micro scale culture chamber | biofilm growth with time | [80] |
| SR-FTIR spectroscopy | circumventing water-absorption barrier | molecular level within biofilms over a long timebiomolecule synthesis during biofilm development | [81] |
| Optical density(LED array & photodiodes) | transparent culture chamber | change in biofilm optical density over the growth period | [82] |
| High-density interdigitated capacitors (µIDES) | dielectric micro-sensors integrated transparent biochip | changes of optical density and impedance caused by biofilm growthdynamic responses of biofilms to shear stress and antimicrobial agent concentration | [83] |
3.2.5. Mimicking Biological Environments
4. Conclusions
Acknowledgments
References
- Zobell, C.E. The effect of solid surfaces upon bacterial activity. J. Bacteriol. 1943, 46, 39–56. [Google Scholar]
- Prince, A.A.; Steiger, J.D.; Khalid, A.N.; Dogrhamji, L.; Reger, C.; Claire, S.E.; Chiu, A.G.; Kennedy, D.W.; Palmer, J.N.; Cohen, N.A. Prevalence of biofilm-forming bacteria in chronic rhinosinusitis. Am. J. Rhinol. 2008, 22, 239–245. [Google Scholar] [CrossRef]
- Nett, J.; Lincoln, L.; Marchillo, K.; Massey, R.; Holoyda, K.; Hoff, B.; VanHandel, M.; Andes, D. Putative role of beta-1,3 glucans in Candida albicans biofilm resistance. Antimicrob. Agents Chemother. 2007, 51, 510–520. [Google Scholar]
- Deligianni, E.; Pattison, S.; Berrar, D.; Ternan, N.G.; Haylock, R.W.; Moore, J.E.; Elborn, S.J.; Dooley, J.S.G. Pseudomonas aeruginosa Cystic Fibrosis isolates of similar RAPD genotype exhibit diversity in biofilm forming ability in vitro. BMC Microbiol. 2010, 10, 38. [Google Scholar] [CrossRef]
- Sawasdidoln, C.; Taweechaisupapong, S.; Sermswan, R.W.; Tattawasart, U.; Tungpradabkul, S.; Wongratanacheewin, S. Growing Burkholderia pseudomallei in biofilm stimulating conditions significantly induces antimicrobial resistance. PLoS One 2010, 5, e9196. [Google Scholar]
- Caliskan, S.; Ozcan, S.K.; Cinar, S.; Corakci, A.; Caliskan, E. In vitro biofilm formation and relationship with antifungal resistance of Candida spp. isolated from vaginal and intrauterine device string samples of women with vaginal complaints. Mikrobiyol. Bul. 2011, 45, 697–706. [Google Scholar]
- Thomas, J.G.; Nakaishi, L.A. Managing the complexity of a dynamic biofilm. J. Am. Dent. Assoc. 2006, 137, 10–15. [Google Scholar]
- Donlan, R.M. Biofilms and device-associated infections. Emerging Infect. Dis. 2001, 7, 277–281. [Google Scholar] [CrossRef]
- Rochex, A.; Godon, J.J.; Bernet, N.; Escudie, R. Role of shear stress on composition, diversity and dynamics of biofilm bacterial communities. Water Res. 2008, 42, 4915–4922. [Google Scholar] [CrossRef]
- Puckett, S.D.; Taylor, E.; Raimondo, T.; Webster, T.J. The relationship between the nanostructure of titanium surfaces and bacterial attachment. Biomaterials 2010, 31, 706–713. [Google Scholar] [CrossRef]
- Dickschat, J.S. Quorum sensing and bacterial biofilms. Nat. Prod. Rep. 2010, 27, 343–369. [Google Scholar] [CrossRef]
- de Kievit, T.R. Quorum sensing in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 2009, 11, 279–288. [Google Scholar] [CrossRef]
- Skolimowski, M.; Nielsen, M.W.; Emneus, J.; Molin, S.; Taboryski, R.; Sternberg, C.; Dufva, M.; Geschke, O. Microfluidic dissolved oxygen gradient generator biochip as a useful tool in bacterial biofilm studies. Lab Chip 2010, 10, 2162–2169. [Google Scholar] [CrossRef]
- Sawyer, L.K.; Hermanowicz, S.W. Detachment of biofilm bacteria due to variations in nutrient supply. Water Sci. Technol. 1998, 37, 211–214. [Google Scholar]
- Jefferson, K.K. What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 2004, 236, 163–173. [Google Scholar]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef]
- Dibdin, G.H.; Assinder, S.J.; Nichols, W.W.; Lambert, P.A. Mathematical model of beta-lactam penetration into a biofilm of Pseudomonas aeruginosa while undergoing simultaneous inactivation by released beta-lactamases. J. Antimicrob. Chemother. 1996, 38, 757–769. [Google Scholar] [CrossRef]
- Mah, T.-F.C.; O’Toole, G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef]
- Shapiro, J.A. Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 1998, 52, 81–104. [Google Scholar] [CrossRef]
- Caldwell, D.E. Post-modern ecology is the environment the organism? Environ. Microbiol. 1999, 1, 279–281. [Google Scholar] [CrossRef]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar]
- Gottenbos, B.; van der Mei, H.C.; Klatter, F.; Nieuwenhuis, P.; Busscher, H.J. In vitro and in vivo antimicrobial activity of covalently coupled quaternary ammonium silane coatings on silicone rubber. Biomaterials 2002, 23, 1417–1423. [Google Scholar] [CrossRef]
- Klausen, M.; Heydorn, A.; Ragas, P.; Lambertsen, L.; Aaes-Jorgensen, A.; Molin, S.; Tolker-Nielsen, T. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 2003, 48, 1511–1524. [Google Scholar] [CrossRef]
- Lecuyer, S.; Rusconi, R.; Shen, Y.; Forsyth, A.; Vlamakis, H.; Kolter, R.; Stone, H.A. Shear stress increases the residence time of adhesion of Pseudomonas aeruginosa. Biophys. J. 2011, 100, 341–350. [Google Scholar]
- Ochoa, J.C.; Coufort, C.; Escudie, R.; Line, A.; Paul, E. Influence of non-uniform distribution of shear stress on aerobic biofilms. Chem. Eng. Sci. 2007, 62, 3672–3684. [Google Scholar] [CrossRef]
- O’Toole, G.A.; Gibbs, K.A.; Hager, P.W.; Phibbs, P.V.; Kolter, R. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 2000, 182, 425–431. [Google Scholar] [CrossRef]
- Davies, D.G.; Chakrabarty, A.M.; Geesey, G.G. Exopolysaccharide production in biofilms—Substratum activation of alginate gene-expression by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 1993, 59, 1181–1186. [Google Scholar]
- Whiteley, M.; Bangera, M.G.; Bumgarner, R.E.; Parsek, M.R.; Teitzel, G.M.; Lory, S.; Greenberg, E.P. Gene expression in Pseudomonas aeruginosa biofilms. Nature 2001, 413, 860–864. [Google Scholar]
- Knobloch, J.K.M.; Nedelmann, M.; Kiel, K.; Bartscht, K.; Horstkotte, M.A.; Dobinsky, S.; Rohde, H.; Mack, D. Establishment of an arbitrary PCR for rapid identification of Tn917 insertion sites in Staphylococcus epidermidis: Characterization of biofilm-negative and nonmucoid mutants. Appl. Environ. Microbiol. 2003, 69, 5812–5818. [Google Scholar]
- Corona-Izquierdo, F.P.; Membrillo-Hernandez, J. A mutation in rpoS enhances biofilm formation in Escherichia coli during exponential phase of growth. FEMS Microbiol. Lett. 2002, 211, 105–110. [Google Scholar] [CrossRef]
- Rachid, S.; Ohlsen, K.; Wallner, U.; Hacker, J.; Hecker, M.; Ziebuhr, W. Alternative transcription factor sigma(B) is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 2000, 182, 6824–6826. [Google Scholar]
- Knobloch, J.K.M.; Bartscht, K.; Sabottke, A.; Rohde, H.; Feucht, H.H.; Mack, D. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: Differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 2001, 183, 2624–2633. [Google Scholar] [CrossRef]
- Yoshida, A.; Kuramitsu, H.K. Multiple Streptococcus mutans genes are involved in biofilm formation. Appl. Environ. Microbiol. 2002, 68, 6283–6291. [Google Scholar] [CrossRef]
- Wen, Z.Z.T.; Burne, R.A. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 2002, 68, 1196–1203. [Google Scholar] [CrossRef]
- Merritt, J.; Qi, F.X.; Goodman, S.D.; Anderson, M.H.; Shi, W.Y. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 2003, 71, 1972–1979. [Google Scholar] [CrossRef]
- Lunsford, R.D.; London, J. Natural genetic transformation in Streptococcus gordonii: comX Imparts spontaneous competence on strain wicky. J. Bacteriol. 1996, 178, 5831–5835. [Google Scholar]
- Kjaergaard, K.; Schembri, M.A.; Ramos, C.; Molin, S.; Klemm, P. Antigen 43 facilitates formation of multispecies biofilms. Environ. Microbiol. 2000, 2, 695–702. [Google Scholar] [CrossRef]
- Toledo-Arana, A.; Valle, J.; Solano, C.; Arrizubieta, M.J.; Cucarella, C.; Lamata, M.; Amorena, B.; Leiva, J.; Penades, J.R.; Lasa, I. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 2001, 67, 4538–4545. [Google Scholar]
- Hufnagel, M.; Koch, S.; Creti, R.; Baldassarri, L.; Huebner, J. A putative sugar-binding transcriptional regulator in a novel gene locus in Enterococcus faecalis contributes to production of Biofilm and prolonged bacteremia in mice. J. Infect. Dis. 2004, 189, 420–430. [Google Scholar] [CrossRef]
- Hussain, M.; Herrmann, M.; von Eiff, C.; Perdreau Remington, F.; Peters, G. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect. Immun. 1997, 65, 519–524. [Google Scholar]
- Gross, M.; Cramton, S.E.; Gotz, F.; Peschel, A. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect. Immun. 2001, 69, 3423–3426. [Google Scholar] [CrossRef]
- Vaudaux, P.E.; Francois, P.; Proctor, R.A.; McDevitt, D.; Foster, T.J.; Albrecht, R.M.; Lew, D.P.; Wabers, H.; Cooper, S.L. Use of adhesion-defective mutants of Staphylococcus aureus to define the role of specific plasma-proteins in promoting bacterial adhesion to canine arteriovenous shunts. Infect. Immun. 1995, 63, 585–590. [Google Scholar]
- Caiazza, N.C.; O’Toole, G.A. Alpha-toxin is required for biofilm formation by Staphylococcus aureus. J. Bacteriol. 2003, 185, 3214–3217. [Google Scholar] [CrossRef]
- Idone, V.; Brendtro, S.; Gillespie, R.; Kocaj, S.; Peterson, E.; Rendi, M.; Warren, W.; Michalek, S.; Krastel, K.; Cvitkovitch, D.; et al. Effect of an orphan response regulator on Streptococcus mutans sucrose-dependent adherence and cariogenesis. Infect. Immun. 2003, 71, 4351–4360. [Google Scholar]
- Heilmann, C.; Schweitzer, O.; Gerke, C.; Vanittanakom, N.; Mack, D.; Götz, F. Molecular basis of intercellular adhesion in the biofilm‐forming Staphylococcus epidermidis. Mol. Microbiol. 1996, 20, 1083–1091. [Google Scholar] [CrossRef]
- Loo, C. Oral streptococcal genes that encode biofilm formation. In Medical Implications of Biofilms; Cambridge University Press: Cambridge, UK, 2003; pp. 189–211. [Google Scholar]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerging Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J. Relevance of microbial extracellular polymeric substances (EPSs). Part I: Structural and ecological aspects. Water Sci. Technol. 2001, 43, 1–8. [Google Scholar]
- Kinnari, T.J.; Esteban, J.; Martin-De-Hijas, N.Z.; Sanchez-Munoz, O.; Sanchez-Salcedo, S.; Colilla, M.; Vallet-Regi, M.; Gomez-Barrena, E. Influence of surface porosity and pH on bacterial adherence to hydroxyapatite and biphasic calcium phosphate bloceramics. J. Med. Microbiol. 2009, 58, 132–137. [Google Scholar] [CrossRef]
- Kapellos, G.E.; Alexiou, T.S.; Payatakes, A.C. Hierarchical simulator of biofilm growth and dynamics in granular porous materials. Adv. Water Resour. 2007, 30, 1648–1667. [Google Scholar] [CrossRef]
- Song, B.; Leff, L.G. Influence of magnesium ions on biofilm formation by Pseudomonas fluorescens. Microbiol. Res. 2006, 161, 355–361. [Google Scholar] [CrossRef]
- Brizzolara, R.A.; Holm, E.R. The effect of solid surface tension and exposure to elevated hydrodynamic shear on Pseudomonas fluorescens biofilms grown on modified titanium surfaces. Biofouling 2006, 22, 431–440. [Google Scholar] [CrossRef]
- Garny, K.; Horn, H.; Neu, T.R. Interaction between biofilm development, structure and detachment in rotating annular reactors. Bioprocess Biosyst. Eng. 2008, 31, 619–629. [Google Scholar] [CrossRef]
- Salek, M.M.; Jones, S.M.; Martinuzzi, R.J. The influence of flow cell geometry related shear stresses on the distribution, structure and susceptibility of Pseudomonas aeruginosa 01 biofilms. Biofouling 2009, 25, 711–725. [Google Scholar] [CrossRef]
- Lee, J.H.; Kaplan, J.B.; Lee, W.Y. Microfluidic devices for studying growth and detachment of Staphylococcus epidermidis biofilms. Biomed. Microdevices 2008, 10, 489–498. [Google Scholar] [CrossRef]
- Tsai, Y.P. Impact of flow velocity on the dynamic behaviour of biofilm bacteria. Biofouling 2005, 21, 267–277. [Google Scholar] [CrossRef]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar]
- Davies, D.G.; Parsek, M.R.; Pearson, J.P.; Iglewski, B.H.; Costerton, J.W.; Greenberg, E.P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 1998, 280, 295–298. [Google Scholar]
- Chopp, D.L.; Kirisits, M.J.; Moran, B.; Parsek, M.R. The dependence of quorum sensing on the depth of a growing biofilm. Bull. Math. Biol. 2003, 65, 1053–1079. [Google Scholar] [CrossRef]
- Shrout, J.D.; Chopp, D.L.; Just, C.L.; Hentzer, M.; Givskov, M.; Parsek, M.R. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 2006, 62, 1264–1277. [Google Scholar] [CrossRef]
- Hunt, S.M.; Werner, E.M.; Huang, B.C.; Hamilton, M.A.; Stewart, P.S. Hypothesis for the role of nutrient starvation in biofilm detachment. Appl. Environ. Microbiol. 2004, 70, 7418–7425. [Google Scholar]
- Applegate, D.H.; Bryers, J.D. Effects of carbon and oxygen limitations and calcium concentrations on biofilm removal processes. Biotechnol. Bioeng. 1991, 37, 17–25. [Google Scholar] [CrossRef]
- Mun, S.M.; Jeong, J.S.; Kim, J.E.; Lee, Y.W.; Yoon, J.Y. Inactivation of Pseudomonas aeruginosa biofilm by dense phase carbon dioxide. Biofouling 2009, 25, 473–479. [Google Scholar] [CrossRef]
- Janakiraman, V.; Englert, D.; Jayaraman, A.; Baskaran, H. Modeling Growth and Quorum Sensing in Biofilms Grown in Microfluidic Chambers. Ann. Biomed. Eng. 2009, 37, 1206–1216. [Google Scholar] [CrossRef]
- Bahar, O.; de la Fuente, L.; Burdman, S. Assessing adhesion, biofilm formation and motility of Acidovorax citrulli using microfluidic flow chambers. FEMS Microbiol. Lett. 2010, 312, 33–39. [Google Scholar] [CrossRef]
- Park, A.; Jeong, H.H.; Lee, J.; Kim, K.P.; Lee, C.S. Effect of shear stress on the formation of bacterial biofilm in a microfluidic channel. BioChip J. 2011, 5, 236–241. [Google Scholar] [CrossRef]
- Mao, H.B.; Cremer, P.S.; Manson, M.D. A sensitive, versatile microfluidic assay for bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 2003, 100, 5449–5454. [Google Scholar]
- Keenan, T.M.; Folch, A. Biomolecular gradients in cell culture systems. Lab Chip 2008, 8, 34–57. [Google Scholar] [CrossRef]
- Long, T.; Ford, R.M. Enhanced transverse migration of bacteria by chemotaxis in a porous T-sensor. Environ. Sci. Technol. 2009, 43, 1546–1552. [Google Scholar] [CrossRef]
- Ahmed, T.; Shimizu, T.S.; Stocker, R. Microfluidics for bacterial chemotaxis. Integr. Biol. 2010, 2, 604–629. [Google Scholar] [CrossRef]
- Diao, J.; Young, L.; Kim, S.; Fogarty, E.A.; Heilman, S.M.; Zhou, P.; Shuler, M.L.; Wu, M.; DeLisa, M.P. A three-channel microfluidic device for generating static linear gradients and its application to the quantitative analysis of bacterial chemotaxis. Lab Chip 2006, 6, 381–388. [Google Scholar] [CrossRef]
- Cheng, S.-Y.; Heilman, S.; Wasserman, M.; Archer, S.; Shuler, M.L.; Wu, M. A hydrogel-based microfluidic device for the studies of directed cell migration. Lab Chip 2007, 7, 763–769. [Google Scholar] [CrossRef]
- Ahmed, T.; Shimizu, T.S.; Stocker, R. Bacterial chemotaxis in linear and nonlinear steady microfluidic gradients. Nano Lett. 2010, 10, 3379–3385. [Google Scholar] [CrossRef]
- Eun, Y.J.; Weibel, D.B. Fabrication of microbial biofilm arrays by geometric control of cell adhesion. Langmuir 2009, 25, 4643–4654. [Google Scholar] [CrossRef]
- Kim, J.; Hegde, M.; Kim, S.H.; Wood, T.K.; Jayaraman, A. A microfluidic device for high throughput bacterial biofilm studies. Lab Chip 2012, 12, 1157–1163. [Google Scholar] [CrossRef]
- Benoit, M.R.; Conant, C.G.; Ionescu-Zanetti, C.; Schwartz, M.; Matin, A. New device for high-throughput viability screening of flow biofilms. Appl. Environ. Microbiol. 2010, 76, 4136–4142. [Google Scholar] [CrossRef]
- Sun, P.; Liu, Y.; Sha, J.; Zhang, Z.Y.; Tu, Q.; Chen, P.; Wang, J.Y. High-throughput microfluidic system for long-term bacterial colony monitoring and antibiotic testing in zero-flow environments. Biosens. Bioelectron. 2011, 26, 1993–1999. [Google Scholar] [CrossRef]
- Kim, J.; Hegde, M.; Jayaraman, A. Co-culture of epithelial cells and bacteria for investigating host-pathogen interactions. Lab Chip 2010, 10, 43–50. [Google Scholar] [CrossRef]
- Kim, K.P.; Kim, Y.G.; Choi, C.H.; Kim, H.E.; Lee, S.H.; Chang, W.S.; Lee, C.S. In situ monitoring of antibiotic susceptibility of bacterial biofilms in a microfluidic device. Lab Chip 2010, 10, 3296–3299. [Google Scholar] [CrossRef]
- Yawata, Y.; Toda, K.; Setoyama, E.; Fukuda, J.; Suzuki, H.; Uchiyama, H.; Nomura, N. Monitoring biofilm development in a microfluidic device using modified confocal reflection microscopy. J. Biosci. Bioeng. 2010, 110, 377–380. [Google Scholar] [CrossRef]
- Holman, H.Y.N.; Miles, R.; Hao, Z.; Wozei, E.; Anderson, L.M.; Yang, H. Real-time chemical imaging of bacterial activity in biofilms using open-channel microfluidics and synchrotron FTIR spectromicroscopy. Anal. Chem. 2009, 81, 8564–8570. [Google Scholar] [CrossRef]
- Meyer, M.T.; Roy, V.; Bentley, W.E.; Ghodssi, R. Development and validation of a microfluidic reactor for biofilm monitoring via optical methods. J. Micromech. Microeng. 2011, 21, 054023. [Google Scholar] [CrossRef]
- Richter, L.; Stepper, C.; Mak, A.; Reinthaler, A.; Heer, R.; Kast, M.; Bruckl, H.; Ertl, P. Development of a microfluidic biochip for online monitoring of fungal biofilm dynamics. Lab Chip 2007, 7, 1723–1731. [Google Scholar] [CrossRef]
- Shumi, W.; Lim, J.; Nam, S.W.; Lee, K.; Kim, S.H.; Kim, M.H.; Cho, K.S.; Park, S. Environmental factors that affect Streptococcus mutans biofilm formation in a microfluidic device mimicking teeth. BioChip J. 2010, 4, 257–263. [Google Scholar] [CrossRef]
- Lee, J.H.; Wang, H.J.; Kaplan, J.B.; Lee, W.Y. Microfluidic approach to create three-dimensional tissue models for biofilm-related infection of orthopaedic implants. Tissue Eng. C Methods 2011, 17, 39–48. [Google Scholar] [CrossRef]
- Boedicker, J.Q.; Vincent, M.E.; Ismagilov, R.F. Microfluidic confinement of single cells of bacteria in small volumes initiates high-density behavior of quorum sensing and growth and reveals its variability. Angew. Chem. Int. Ed. Engl. 2009, 48, 5908–5911. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kim, J.; Park, H.-D.; Chung, S. Microfluidic Approaches to Bacterial Biofilm Formation. Molecules 2012, 17, 9818-9834. https://doi.org/10.3390/molecules17089818
Kim J, Park H-D, Chung S. Microfluidic Approaches to Bacterial Biofilm Formation. Molecules. 2012; 17(8):9818-9834. https://doi.org/10.3390/molecules17089818
Chicago/Turabian StyleKim, Junghyun, Hee-Deung Park, and Seok Chung. 2012. "Microfluidic Approaches to Bacterial Biofilm Formation" Molecules 17, no. 8: 9818-9834. https://doi.org/10.3390/molecules17089818
APA StyleKim, J., Park, H.-D., & Chung, S. (2012). Microfluidic Approaches to Bacterial Biofilm Formation. Molecules, 17(8), 9818-9834. https://doi.org/10.3390/molecules17089818