Synthesis and Characterization of Some New Cu(II), Ni(II) and Zn(II) Complexes with Salicylidene Thiosemicarbazones: Antibacterial, Antifungal and in Vitro Antileukemia Activity
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry

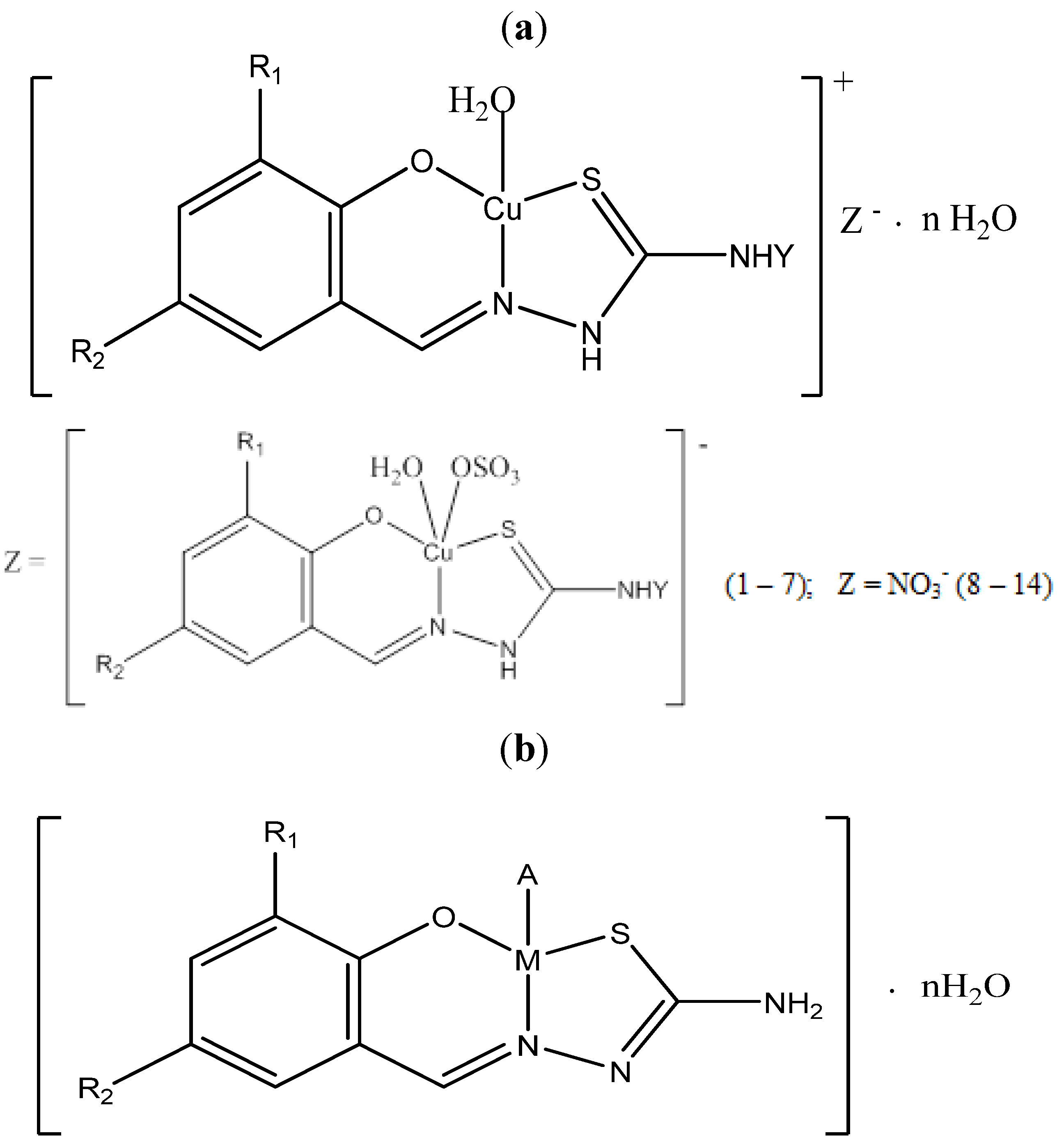
| Comp. No. | Molecular formula | Mr b | µ effc B.M. | C, H, N, calc (found) % | M(3d) d % | IR (cm−1) | η, % e | T, C f dec |
|---|---|---|---|---|---|---|---|---|
| 1 | C16H24Cu2N6O10S3 [Cu(H2O)(HL1)][Cu(H2O)(HL1)SO4] . 2H2O | 684 | 2.14 | C: 28.1(28.5); H: 3.5 (3.0); N: 12.3(12.5); S: 14.0 (13.7) | 18.7 (18.6) | H2O (3585, 1575, 920); NH2 (3435, 3420); NH(3335, 3220, 3145); C= N (1605); C-O (1200); C = S (781); Cu-N (510, 415); Cu-O (470); Cu-S (450) | 65 | 460 |
| 2 | C28H30Cu2N6O9S3 [Cu(H2O)(HL8)][Cu(H2O)][(HL8)SO4] . H2O | 818 | 2.07 | C: 41.1(41.4); H: 3.7 (3.4); N: 10.3(10.3); S: 11.7 (11.6) | 15.6 (15.8) | H2O (3580, 1570, 925); NH (3325, 3222, 3143); C = N (1600); C-0(1195); C = S (780); Cu-N(517, 428); Cu-O (472); Cu-S (445) | 64 | 450 |
| 3 | C16H22Cu2N8O14S3 [Cu(H2O)(HL4)][Cu(H2O)(HL4)(SO4) ] . H2O | 774 | 1.98 | C: 24.8 (24.5); H: 2.8(2.7); N: 14.5 (14.8); S: 12.4 (12.7) | 16.5 (16.3) | H2O (3575, 1570, 922); NH2 (3445, 3425); NH (3330 3230, 3140); C = N (1590); C-O (1195); C = S (776); Cu-N (530, 410); Cu-O (470); Cu-S (465) | 77 | 425 |
| 4 | C28H30Cu2N8O14S3 [Cu(H2O)(HL1°)][Cu(H2O)(HL1°)(SO4)]. 2H2O | 926 | 2.09 | C: 36.3 (36.5); H: 3.2 (3.0); N: 12.1 (12.4); S: 10.4 (10.1) | 13.8 (14.1) | H2O (3580, 1583, 915); NH (3315, 3230, 3138); C = N (1585); C-O (1197); C = S (779); Cu-N (525, 430); Cu-O (475); Cu-S(440) | 72 | 410 |
| 5 | C16H26Br2Cu2N6O12S3 [Cu(H2O)(HL3)][Cu(H2O)(HL3)(SO4)] . 4H2O | 878 | 1.85 | C: 21.9 (22.2); H: 3.3 (3.3); Br: 18.2 (18.4); N: 9.6 (9.4); S: 10.3 (10.5) | 14.6 (14.4) | H2O (3565, 1575, 935); NH2 (3445, 3430); NH (3340, 3230, 3137); C = N (1590); C-O (1205); C = S (780); Cu-N (505, 430); Cu-O (485); Cu-S (462) | 69 | 450 |
| 6 | C28H28Br2Cu2N6O9S3 [Cu(H2O)(HL9)][Cu(H2O)(HL9)(SO4)] . H2O | 976 | 1.91 | C: 34.4 (34.0); H: 2.9 (2.7); Br: 16.4 (16.5); N: 8.6 (8.4); S: 9.8 (9.9) | 13.1 (12.8) | H2O (3580, 1565, 930); NH (3330, 3225, 3145); C = N (1585); C-O (1203); C = S (778); Cu-N (525, 425); Cu-O (484); Cu-S (465) | 56 | 435 |
| 7 | C16H20Cl2Cu2N6O9S3 [Cu(H2O)(HL2)][Cu(H2O)(HL2)(SO4)] . H2O | 735 | 1.79 | C: 26.1 (26.3); H: 2.7 (2.4); Cl: 9.7 (10.0); N: 11.4 (11.5); S: 13.1 (13.3) | 17.4 (17.7) | H2O (3585, 1575, 920); NH2 (3430, 3430); NH (3335, 3220, 3145); C = N (1595); C-0(1200); C = S(785); Cu-N (528, 410); Cu-O (482); Cu-S (464) | 78 | 430 |
| 8 | C8H12CuN4O6S [Cu(H2O) (HL1)]NO3. H2O | 356 | 1.87 | C: 27.0 (27.3); H: 3.4 (3.1); N: 15.7 (15.5); S: 9.0 (9.4) | 18.0 (18.2) | H2O (3580, 1574, 915); NH2 (3440, 3430); NH (3325, 3230, 3140); C = N(1600); C-0(1200); C = S(776); Cu-N(530, 410); Cu-O(480); Cu-S(450) | 70 | 390 |
| 9 | C14H16CuN4O6S [Cu (H2O)(HL8)]NO3. H2O | 432 | 2.12 | C: 38.9 (38.4); H: 3.7 (3.5); N: 13.0 (13.1); S: 7.4 (7.2) | 14.8 (14.5) | H2O (3576, 1570, 930); NH(3345, 3227, 3146); C = N(1595); C-O (1198); C = S (787); Cu-N (525, 430); Cu-O (465); Cu-S (440) | 54 | 380 |
| 10 | C8H11CuN5O8S [Cu (H2O)(HL4)]NO3. H2O | 401 | 1.85 | C: 23.9 (24.2); H: 2.7 (2.5); N: 17.5 (17.1); S: 8.0 (8.3) | 16.0 (16.3) | H2O (3570, 1565, 925); NH2 (3445, 3430); NH (3325, 3215, 3140); C = N (1598); C-O (1195); C = S (777); Cu-N (525, 410); Cu-O (475); Cu-S (440) | 76 | 325 |
| 11 | C14H17CuN5O9S [Cu (H2O)(HL1°)]NO3. 2H2O | 495 | 1.94 | C: 33.9 (34.1); H: 3.4 (3.5); N: 14.1 (14.2); S: 6.5 (6.9) | 12.9 (12.7) | H2O (3590, 1585, 915); NH(3325, 3225, 3140); C = N(1593); C-0(1192); C = S(783); Cu-N(525, 430); Cu-O(480); Cu-S(455) | 80 | 315 |
| 12 | C8H11BrCuN4O6S [Cu (H2O)(HL3)]NO3. H2O | 435 | 1.80 | C: 22.1 (21.8); H: 2.5 (2.2); N: 12.9 (13.2); S: 7.4 (7.5) | 14.7 (14.5) | H2O (3585, 1575, 920); NH2(3430, 3415); NH(3335, 3220, 3145); C = N(1595); C-0(1195); C = S(784); Cu-N(525, 425); Cu-O(475); Cu-S(460) | 52 | 370 |
| 13 | C14H19BrCuN4O8S [Cu(H2O)(HL9)]NO3. 3H2O | 547 | 1.97 | C: 30.7 (30.9); H: 3.4 (3.2); Br: 14.6 (14.4); N: 10.2 (9.9); S: 5.8 (5.7) | 11.7 (11.5) | H2O (3570, 1565, 925); NH(3330, 3210, 3135); C = N(1590); C-0(1197); C = S(780); Cu-N(530, 423); Cu-O(470); Cu-S(465) | 65 | 360 |
| 14 | C8H11ClCuN4O6S [Cu(H2O)(HL2)]NO3. H2O | 390.5 | 2.03 | C: 24.6 (24.6); H: 2.8 (2.5); Cl: 9.1 (9.3); N: 14.3 (14.5); S: 8.2 (8.5) | 16.4 (16.1) | H2O (3585, 1575, 920); NH2(3435, 3425); NH(3335, 3220, 3145); C = N(1605); C-0(1193); C = S(780); Cu-N(515, 430); Cu-O(490); Cu-S(455) | 75 | 365 |
| 15 | C13H12CuN4OS [Cu L1Py] | 336 | 1.78 | C: 46.4 (46.5); H: 3.6 (3.5); N: 16.7 (16.5); S: 9.5 (9.4) | 19.0 (18.8) | NH2(3440, 3425); C = N(1590, 1580, 1575, 1310); C-0 (1215); C-S ( 760); Cu-N(530, 425); Cu-O(475); Cu-S(460) | 71 | 460 |
| 16 | C13H11ClCuN4OS [Cu L2Py] | 370.5 | 1.78 | C: 42.1 (42.0); H: 3.0 (2.9); Cl: 9.6 (9.5); N: 15.1 (15.0); S: 8.6 (8.4) | 17.3 (17.0) | NH2 (3435, 3420); C = N (1585, 1580, 1570, 1305); C-O (1225); C-S (750); Cu-N (510, 405); Cu-O (470); Cu-S (465) | 72 | 440 |
| 17 | C13H11BrCuN4OS [Cu L3Py] | 415 | 1.93 | C: 37.6 (37.5); H: 2.7 (2.5); Br: 19.3 (19.0); N: 13.5 (13.3); S: 7.7 (7.5) | 15.4 (15.5) | NH2 (3430, 3420); C = N (1585, 1580, 1575, 1300); C-O (1210); C-S (750); Cu-N (515, 410); Cu-O (475); Cu-S (465) | 75 | 450 |
| 18 | C13H11CuN5O3S [Cu L4Py] | 381 | 1.84 | C: 40.9 (40.8); H: 2.9 (2.8); N: 18.4 (18.2); S: 8.4 (8.3) | 16.8 (16.7) | NH2 (3440, 3425); C = N (1585, 1580, 1570, 1315); C-O (1220); C-S ( 770); Cu-N (525, 410); Cu-O (470); Cu-S (465) | 69 | 400 |
| 19 | C14H14CuN4OS [Cu L5Py] | 350 | 1.75 | C: 48.0 (47.8); H: 4.0 (3.9); N: 16.0 (15.9); S: 9.1 (9.0) | 18.3 (18.0) | NH2 (3430, 3425); C = N (1590, 1585, 1570, 1315); C-O (1220); C-S ( 770); Cu-N (520, 415); Cu-O (470); Cu-S (465) | 70 | 460 |
| 20 | C13H10Cl2CuN4OS [Cu L6Py] | 405 | 1.80 | C: 38.5 (38.3); H: 2.5 (2.4); Cl: 17.5 (17.4); N: 13.8 (13.6); S: 7.9 (7.8) | 15.8 (15.7) | NH2 (3435, 3425); C = N (1585, 1580, 1575, 1305); C-O (1205); C-S ( 770); Cu-N (515, 430); Cu-O (485); Cu-S (47 0) | 76 | 410 |
| 21 | C8H12Br2CuN4O3S [Cu L7(NH3)] × 2H2O | 468 | 1.87 | C: 20.5 (20.4); H: 2.6 (2.5); Br: 34.2 (34.0); S: 6.8(6.7) | 11.9(1.7) | NH2 (3440, 3425); NH (3330, 3215, 3150); C = N (1582, 1585); C-O (1225); C-S (748); Cu-N (540, 425); Cu-O (490); Cu-S (410); | 78 | 310 |
| 22 | C14H14 Br2CuN4O2S [Cu L7(4-MePy)] × H2O | 526 | 1.79 | C: 31.9 (31.8); H: 2.7 (2.5); Br: 31.5(31.3); S: 6.3(6.0) | 12.2(12.3) | NH2 (3435,3430); C = N (1580,1585); C-O (1225); CNC (1042); C-S (748); Cu-O (540); Cu-O (490); Cu-S (410) | 77 | 380 |
| 23 | C14H12 Br2CuN4OS [Cu L7(3-MePy)] | 508 | 1.99 | C: 33.1 (33.0); H: 2.4 (2.2); Br: 31.5(31.4); S: 6.3(6.1) | 12.6(12.4) | NH2 (3440,3425); C = N (1580,1585); C-O (1225); CNC (1042); C-S (748); Cu-O (540); Cu-O (490); Cu-S (410) | 76 | 390 |
| 24 | C14H12 Br2CuN4OS [Cu L7(2-MePy)] | 508 | 1.92 | C: 33.1 (32.9); H: 2.4 (2.5); Br: 31.5(31.4); S: 6.3(6.0) | 12.6(12.3) | NH2 (3440,3430); C = N (1580,1585); C-O (1225); CNC (1042); C-S (748); Cu-O (540); Cu-O (490); Cu-S (410) | 71 | 345 |
| 25 | C18H17Br2CuN7O3S3 [Cu L7(Etz)] | 699 | 1.35 | C: 30.9 (31.0); H: 2.4 (2.2); Br: 22.9 (22.7); N: 14.0(13.9); S: 13.7(13.5) | 9.2(8.6) | NH2 (3435,3425, 3420, 3410); C = N (1610, 1600, 1585); SO2 (1320, 1140), C-O (1215); Cu-N (540, 415); Cu-O (490); Cu-S (440) | 81 | 470 |
| 26 | C14H13Br2CuN5O3S2 [Cu L7(Str)] | 587 | 1.28 | C: 28.6 (28.5); H: 2.2 (2.0); Br: 27.3 (27.0); N: 11.9 (12.0); S: 10.9 (10.7) | 10.9 (11.0) | NH2 (3415,3420,3405,3415); C = N (1600, 1585); SO2 (1325, 1140); C-O (1210); Cu-N (525, 410); Cu-O (475); Cu-S (455) | 68 | 430 |
| 27 | C16H15Br2CuN5O4S2 [Cu L7(Sfc)] | 629 | 1.31 | C: 30.5 (30.3); H 2.4 (2.5); Br: 25.4 (25.3); N: 11.1 (11.0); S: 10.2 (10.0) | 10.2 (10.1) | NH2 (3420,3415,3415,3405); C = N (1605, 1590); SO2 (1320, 1145); C-O (1215); Cu-N (530, 425); Cu-O (480); Cu-S (465); | 63 | 450 |
| 28 | C17H14Br2CuN6O3S3 [Cu L7(Nor)] | 670 | 1.35 | C: 30.4 (30.2); H: 2.1 (2.0); Br: 23.9 (24.0); N: 12.5 (12.3); S: 14.3 (14.2) | 9.6 (9.5) | NH2 (3430, 3425, 3415, 3410); C = N (1610, 1605, 1590); SO2 (1315, 1145); C-O (1210); Cu-N (530, 420); Cu-O (480); Cu-S (455); | 69 | 470 |
| 29 | C20H19Br2CuN7O3S2 [Cu L7(Sdm)] | 693 | 1.22 | C: 34.6 (34.5); H: 2.7 (2.5); Br: 23.1 (23.0); N: 14.1 (14.0); S: 9.2 (9.0) | 9.2 (9.1) | NH2 (3440, 3430, 3425, 3415); C = N (1610, 1600, 1595); SO2 (1310, 1150); C-O (1215); Cu-N (510, 425); Cu-O (475); Cu-S (450); | 68 | 460 |
| 30 | C18H19CuN7O3S3 [Cu L1(Etz)] | 541 | 1.45 | C: 39.9 (40.0); H: 3.5 (3.4); N: 18.1 (17.9); S: 17.7(17.5) | 11.8(11.6) | NH2 (3435, 3430, 3425, 3415); C = N (1600, 1595, 1590); SO2 (1310, 1140); C-O (1225); Cu-N (515, 410); Cu-O (470); Cu-S (465) | 70 | 500 |
| 31 | C18H23NiN7O5S3 [Ni L1(Etz)] × 2H2O | 572 | dia | C: 37.8 (37.5); H: 4.0 (3.8); N: 17,1 (17.0); S: 16.8 (16.6) | 10.3(10.2) | NH2 (3430, 3430, 3420, 3410); C = N (1605, 1595, 1590); SO2 (1315, 1145); C-O (1220); Cu-N (525, 415); Cu-O (475); Cu-S (460) | 80 | 380 |
| 32 | C18H19N7O3S3Zn [Zn L1(Etz)] | 542 | dia | C: 39.9 (40.0); H: 3.5 (3.4); N: 18.1 (18.0); S: 17.7 (17.5) | 12.0 (11.8) | NH2 (3430, 3430, 3420, 3415); C = N (1605, 1595, 1585); SO2 (1315, 1140); C-0 (1215); Cu-N (525, 425); Cu-O (480); Cu-S (470) | 75 | 490 |
2.1.1. X-ray Structure of [Cu(H2O)(HL3)][Cu(H2O)(HL3)(SO4)]·4H2O (5)
2.1.2. IR Spectra and Coordination Mode
2.1.3. Magnetochemistry
2.1.4. Thermal Decomposition
2.1.5. NMR Spectra
2.1.6. Mass Spectra
| Molecular formula | Mw
(g/mol) | Molecular ion peak [M]+ | The peaks due to complex fragmentation | |||
|---|---|---|---|---|---|---|
| [Cu(H2O)(HL1)][Cu(H2O)(HL1)SO4] . 2H2O (1) | 684 | 274.8 | 101.2 | 170.3 | 203.4 | |
| [Cu(H2O)(HL4)][Cu(H2O)(HL4)(SO4)] . H2O (3) | 774 | 319.7 | 147.3 | 216.5 | 296.3 | |
| [Cu(H2O)(HL2)][Cu(H2O)(HL2)(SO4)] . H2O (7) | 735 | 309.6 | 136.7 | 206.3 | 287.5 | |
| [Cu (H2O)(HL8)]NO3 . H2O (9) | 432 | 350.9 | 101.7 | 171.4 | 203.8 | 320.2 |
| [Cu (H2O)(HL1°)]NO3 . 2H2O (11) | 495 | 395.6 | 147.7 | 220.2 | 286.3 | 372.1 |
| [Cu(H2O)(HL9)]NO3 . 3H2O (13) | 547 | 429.8 | 181.2 | 229.1 | 295.2 | 398.8 |
| [Cu L2Py] (16) | 370.5 | 369.8 | 136.7 | 207.5 | 292.1 | 322.6 |
| [Cu L5Py] (19) | 350 | 349.3 | 132.1 | 203.3 | 289.2 | 318.5 |
| [Cu L7(4-MePy)] × H2O (22) | 526 | 506.8 | 262.3 | 327.8 | 403.2 | 498.8 |
| [Cu L7(Etz)] (25) | 699 | 698.2 | 296.3 | 357.5 | 434.4 | 544.2 |
| [Cu L7(Str)] (26) | 587 | 586.1 | 284.1 | 345.6 | 422.1 | 532.4 |
| [Ni L1(Etz)] × 2H2O (31) | 572 | 536 | 269.5 | 330.6 | 401.3 | 517.8 |
| [Zn L1(Etz)] (32) | 542 | 541.4 | 282.2 | 344.2 | 416.1 | 527.4 |
2.2. Biological Activity
2.2.1. Antiproliferative Activity of Human Leukemia HL-60 Cells
| Schiff base | (X)N-NH-C(S)-NH(Y) | Inhibition of cell proliferation (%) | ||||
|---|---|---|---|---|---|---|
| X | Y | 10 µM | 1 µM | 0.1µM | ||
 | ||||||
| R1 | R2 | |||||
| H2L1 | H | H | H | 20 | 10 | 0 |
| H2L2 | H | Cl | H | 0 | 0 | 0 |
| H2L3 | H | Br | H | 5 | 0 | 0 |
| H2L4 | H | NO2 | H | 0 | 0 | 0 |
| H2L5 | H | CH3 | H | 5 | 0 | 0 |
| H2L6 | Cl | Cl | H | 10 | 0 | 0 |
| H2L7 | Br | Br | H | 0 | 0 | 0 |
| H2L8 | H | H | C6H5 | 90 | 0 | 0 |
| H2L9 | H | Br | C6H5 | 75 | 0 | 0 |
| H2L10 | H | NO2 | C6H5 | 70 | 0 | 0 |
| Complex a | Structural formula of copper complex | Inhibition of cell proliferation (%) b | Complex a | Structural formula of metal complexes | Inhibition of cell proliferation (%)b | ||||||||
 | 10 µM | 1 µM | 0.1 µM |  | 10 µM | 1 µM | 0.1 µM | ||||||
| R1 | R2 | Y | R1 | R2 | A | ||||||||
| 1 | H | H | H | 98 | 50 | 0 | 15 | H | H | Py | - | 35 | 10 |
| 2 | H | H | -C6H5 | 100 | 90 | 0 | 16 | H | Cl | Py | - | 25 | 5 |
| 3 | H | NO2 | H | 90 | 70 | 0 | 17 | H | Br | Py | - | 50 | 0 |
| 4 | H | NO2 | -C6H5 | 96 | 78 | 0 | 18 | H | NO2 | Py | - | 10 | 0 |
| 5 | H | Br | H | 95 | 90 | 0 | 19 | H | CH3 | Py | - | 55 | 0 |
| 6 | H | Br | -C6H5 | 90 | 90 | 0 | 20 | Cl | Cl | Py | - | 60 | 10 |
| 7 | H | Cl | H | 95 | 95 | 0 | 21 | Br | Br | NH3 | - | 25 | 0 |
| 8 | H | H | H | 100 | 95 | 0 | 22 | Br | Br | 4-MePy | - | 20 | 0 |
| 9 | H | H | -C6H5 | 100 | 100 | 0 | 23 | Br | Br | 3-MePy | - | 30 | 15 |
| 10 | H | NO2 | H | 100 | 90 | 0 | 24 | Br | Br | 2-MePy | - | 30 | 5 |
| 11 | H | NO2 | -C6H5 | 100 | 90 | 0 | 25 | Br | Br | Ethazole | - | 60 | 15 |
| 12 | H | Br | H | 98 | 95 | 0 | 26 | Br | Br | Streptocide | 65 | 40 | 5 |
| 13 | H | Br | C6H5 | 100 | 80 | 0 | 27 | Br | Br | Sulfocile | 65 | 40 | 5 |
| 14 | H | Cl | H | 100 | 90 | 0 | 28 | Br | Br | Norsulfosole | 65 | 55 | 5 |
| 29 | Br | Br | Sulfadimizine | 65 | 40 | 5 | |||||||
| DOX | 100 | 100 | 30 | 30 | H | H | Ethazole | 60 | 65 | 0 | |||
| 31 | H | H | Ethazole | 5 | 5 | 5 | |||||||
| 32 | H | H | Ethazole | 10 | 5 | 0 | |||||||
2.2.2. Antibacterial Activity
| Stem | Complexes c | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 21 | 22 | 23 | 24 | 25 | 30 | Furacillinum | Nystatin | |||
| Staphylococcus aureus | Wood-46 | MIC | 0.29 | 0.145 | 0.145 | 0.29 | 0.06 | 0.03 | 9.35 | - |
| MBC | 0.29 | 0.145 | 0.145 | 0.29 | 0.06 | 0.03 | 9.35 | - | ||
| Smith | MIC | 0.29 | 0.145 | 0.29 | 0.29 | - | - | 9.35 | - | |
| MBC | 0.58 | 0.29 | 0.29 | 0.58 | - | - | 9.35 | - | ||
| 209-P | MIC | 0.58 | 0.29 | 0.29 | 0.29 | 0.06 | 0.03 | 18.7 | - | |
| MBC | 0.58 | 1.16 | 1.16 | 0.58 | 0.06 | 0.03 | 18.7 | - | ||
| Staphylococcus saprophyticus | MIC | 0.29 | 0.29 | 0.145 | 0.29 | 0.12 | 0.03 | 9.35 | - | |
| MBC | 0.29 | 0.58 | 0.145 | 0.29 | 0.24 | 0.06 | 18.7 | - | ||
| Streptococcus( group A) | MIC | 0.036 | 0.009 | 1.16 | 0.29 | 0.12 | 0.06 | - | - | |
| MBC | 0.072 | 0.036 | 2.33 | 0.58 | 0.24 | 0.06 | - | - | ||
| Enterococcus faecalis | MIC | - | - | - | - | 0.06 | 0.03 | 37.5 | - | |
| MBC | - | - | - | - | 0.06 | 0.097 | 37.5 | - | ||
| Escherichia coli, O-111 | MIC | 1.16 | 9.35 | 18.7 | 4.67 | 15.6 | 15.6 | 18.7 | - | |
| MBC | 37.5 | 9.35 | 18.7 | 9.35 | 31.2 | 15.6 | 37.5 | - | ||
| Salmonella typhimurium | MIC | 2.33 | 4.67 | 4.67 | 0.29 | 1.95 | 7.8 | 75 | - | |
| MBC | 9.35 | 4.67 | 1000 | 75 | 62.5 | 31.2 | 150 | - | ||
| Salmonella enteritidis | MIC | 2.33 | 9.35 | 4.67 | 1.16 | - | - | 9.35 | - | |
| MBC | 600 | 9.35 | 2000 | 300 | - | - | 9.35 | - | ||
| Klebsiella pneumoniae | MIC | 0.58 | 1.16 | 0.29 | 0.29 | 1.95 | 7.8 | >300 | - | |
| MBC | 600 | 300 | 400 | 300 | 62.5 | 15.6 | >300 | - | ||
| Pseudomonas aeruginosa | MIC | 2000 | 1000 | 2000 | >4000 | 1000 | 250 | >300 | - | |
| MBC | >4000 | 1000 | 4000 | >4000 | >4000 | 250 | >300 | - | ||
| Proteus vulgaris | MIC | 0.29 | 1000 | 1.16 | 1.16 | 0.49 | 7.8 | 150 | - | |
| MBC | 2000 | >4000 | 4000 | 150 | 7.8 | 15.6 | 300 | - | ||
| Proteus mirabilis | MIC | 2.33 | 1000 | 9.35 | 1.16 | - | - | 150 | - | |
| MBC | 1000 | >4000 | >4000 | >4000 | - | - | 300 | - | ||
| Aspergillus niger | MIC | - | 150 | 9.3 | 18.7 | - | - | - | 240 | |
| MBC | - | 150 | 9.3 | 18.7 | - | - | - | 240 | ||
| Aspergillus fumigatus | MIC | - | 300 | 300 | 300 | - | - | - | 240 | |
| MBC | - | 300 | 300 | 300 | - | - | - | 240 | ||
| Candida albicans | MIC | - | 37.5 | 37.5 | 37.5 | - | - | - | 80 | |
| MBC | - | 37.5 | 37.5 | 37.5 | - | - | - | 80 | ||
| Penicillium | MIC | - | 18.7 | 37.5 | 37.5 | - | - | - | 80 | |
| MBC | - | 18.7 | 37.5 | 37.5 | - | - | - | 80 | ||
| LD50, mg/kg | - | - | - | 1420 | - | 4250 | 166.7 | - | ||
3. Experimental
3.1. Chemistry
3.1.1. General Procedures for the Synthesis of the Schiff Bases H2L1–H2L10
3.1.2. General Procedure for the Preparation of Complexes 1–32
3.2. Cytotoxicity Assay
3.2.1. Preparation of Test Solutions
3.2.2. Cell Culture
3.2.3. Cell Proliferation Assay
3.3. Antibacterial Activity
3.4. Antifungal Bioassay
4. Conclusions
Supplementary Data
Acknowledgments
Conflicts of Interest
References
- Rosu, T.; Pahontu, E.; Pasculescu, S.; Georgescu, R.; Stanica, N.; Curaj, A.; Popescu, A.; Leabu, M. Synthesis, characterization antibacterial and antiproliferative activity of novel Cu(II) and Pd(II) complexes with 2-hydroxy-8-R-tricyclo[7.3.1.0.2,7] tridecane-13-one thiosemicarbazone. Eur. J. Med. Chem. 2010, 45, 1627–1634. [Google Scholar] [CrossRef]
- Leovac, V.M.; Bogdanović, G.A.; Jovanović, L.S.; Joksović, L.; Marcović, V.; Joksovic, M.D.; Denčić, S.M.; Isaković, A.; Marković, I.; Heinemann, F.W.; et al. Snthesis, characterization and antitumor activity of polymeric copper (II) complexes with thiosemicarbazones of 3-methyl-5-oxo-1-phenyl-3-pyrazolin-4-carboxaldehyde and 5-oxo-3-phenyl-3-pyrazolin-4-carboxaldehyde. J. Inorg. Biochem. 2011, 105, 1413–1421. [Google Scholar] [CrossRef]
- Hanafy, A.I.; Maki, A.B.K.T.; Mostafa, M.M. Synthesis, structural and catalytic activity of binaphthyl macrocyclic complexes. Transition Met. Chem. 2007, 32, 960–966. [Google Scholar] [CrossRef]
- Fernandez, M.C.; Basitida, R.; Macias, A.; Valencia, L.; Lourida, P.P. ifferent nuclearities of M(II) nitrate complexes (M=Co, Ni, Cu and Cd) with a tetrapyridyl pendant-armed hexaazamacrocyclic ligand. Polyhedron 2006, 25, 783–792. [Google Scholar] [CrossRef]
- Ilhan, S.; Temel, H. Synthesis and spectral studies of macrocyclic Pb(II), Zn(II), Cd(II) and La(III) complexes derived from 1,4-bis(3-aminopropoxy)butane with metal nitrate and salicylaldehyde derivatives. Transition Met Chem. 2007, 32, 1039–1046. [Google Scholar]
- Casas, J.S.; García-Tasende, M.S.; Sánchez, A.; Sordo, J.; Touceda, A. Coordination modes of 5-pyrazolones: A solid-state overview. Coord. Chem. Rev. 2007, 251, 1561. [Google Scholar] [CrossRef]
- Lindoy, L.F. The Chemistry of Macrocyclic Ligand Complexes; Cambridge University Press: Cambridge, UK, 1989. [Google Scholar]
- Ali, M.A.; Mirza, A.H.; Abu Bakar, H.J.H.; Bernhardt, P.V. Preparation and structural characterization of nickel(II), cobalt(II), zinc(II) and tin(IV) complexes of the isatin Schiff bases of S-methyl and S-benzyldithiocarbazates. Polyhedron 2011, 30, 556–564. [Google Scholar] [CrossRef]
- Youssef, N.S.; El-Seidy, A.M.A.; Schiavoni, M.; Castano, B.; Ragaini, F.; Gallo, E.; Caselli, A. Thiosemicarbazone copper complexes as competent catalysts for olefin cyclopropanations. J. Organometallic Chem. 2012. [Google Scholar] [CrossRef]
- Pathan, A.H.; Bakale, R.P.; Naik, G.N.; Frampton, C.S. Synthesis, crystal structure, redox behavior and comprehensive studies on DNA binding and cleavage properties of transition metal complexes of a fluoro substituted thiosemicarbazone derived from ethyl pyruvate. Polyhedron 2012, 34, 149–156. [Google Scholar] [CrossRef]
- Saif, M.; Mashaly, M.M.; Eid, M.F.; Fouad, R. Synthesis, characterization and thermal studies of binary and/or mixed ligand complexes of Cd(II), Cu(II), Ni(II) and Co(III) based on 2-(Hydroxybenzylidene) thiosemicarbazone: DNA binding affinity of binary Cu(II) complex. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2012, 92, 347–356. [Google Scholar] [CrossRef]
- Chandra, S.; Gupta, N.; Gupta, R.; Bawa, S.S. Biologically Relevent Macrocylic Complexes of Copper Spectral, Magnetic Thermal and Antibactrial Approach. Transition Met Chem. 2006, 31, 147–151. [Google Scholar] [CrossRef]
- Tan, K.W.; Seng, H.L.; Lim, F.S.; Cheah, Sh.-Ch.; Ng, C.H.; Koo, K.S.; Mustafa, M.R.; Ng, S.W.; Maah, M.J. Towards a selective cytotoxic agent for prostate cancer: Interaction of zinc complexes of polyhydroxybenzaldehyde thiosemicarbazones with topoisomerase. Polyhedron 2012, 38, 275–284. [Google Scholar] [CrossRef]
- Raja, D.S.; Bhuvanesh, N.S.P.; Natarajan, K. Biological evaluation of a novel water soluble sulphur bridged binuclear copper(II) thiosemicarbazone complex. Eur. J. Med. Chem. 2011, 46, 4584–4594. [Google Scholar] [CrossRef]
- Ramachandran, E.; Thomas, S.P.; Poornima, P.; Kalaivani, P.; Prabhakaran, R.; Padma, V.V.; Natarajan, K. Evaluation of DNA binding, antioxidant and cytotoxic activity of mononuclear Co(III) complexes of 2-oxo-1,2-dihydrobenzoh]quinoline-3-carbaldehyde thiosemicarbazones. Eur. J. Med. Chem. 2012, 50, 405–415. [Google Scholar] [CrossRef]
- Chohan, Z.H.; Pervez, H.; Rauf, A.; Khan, K.M.; Supuran, C.T. Antibacterial cobalt (II), copper (II), nickel (II) and zinc (II) complexes of mercaptothiadiazole--derived furanyl, thienyl, pyrrolyl, salicylyl and pyridinyl Schiff bases. J. Enz. Inh. Med. Chem. 2006, 21, 193. [Google Scholar] [CrossRef]
- Hall, I.H.; Lackey, C.B.; Kistler, T.D.; Durham,, R.W., Jr.; Jouad, E.M.; Khan, M.; Thanh, X.D.; Djebbar-Sid, S.; Benali-Baitich, O.; Bouet, G.M. Cytotoxicity of copper and cobalt complexes of furfural semicarbazone and thiosemicarbazone derivatives in murine and human tumor cell lines. Pharmazie 2000, 937–941. [Google Scholar]
- Baldini, M.; Belicchi-Ferrari, M.; Bisceglie, F.; Dall’Aglio, P.P.; Pelosi, G.; Pinelli, S.; Tarasconi, P. Copper (II) complexes with substituted thiosemicarbazones of α-ketoglutaric acid: synthesis, X-ray structures, DNA binding studies, and nuclease and biological activity. Inorg. Chem. 2004, 43, 7170–7179. [Google Scholar] [CrossRef]
- Belicchi, F.M.; Bisceglie, F.; Pelosi, G.; Tarasconi, P.; Albertini, R.; Dall’Aglio, P.P.; Pinelli, S.; Bergamo, A.; Sava, G.A.J. Synthesis, characterization and biological acvity of copper complexes with pyridoxal thiosemicarbazone derivatives. X-ray crystal structure of three dimeric complexes. Inorg. Biochem. 2004, 98, 301–312. [Google Scholar] [CrossRef]
- Gulea, A.; Poirier, D.; Roy, J.; Stavila, V.; Bulimestru, I.; Ţapcov, V.; Bârcă, V.M.; Popovschi, L. In vitro antileukemia, antibacterial and antifungal activities of some 3d metal complexes: Chemical synthesis and structure–activity relationships. J. Enz. Inh. Med. Chem. 2008, 23, 806–818. [Google Scholar] [CrossRef]
- Gulea, A.P.; Samusi, N.M.; Prisacari, V.I.; Ţapcov, V.I.; Buraciov, S.A.; Spinu, S.N.; Bejenari, N.P.; Poirier, D.; Roy, J. Synthesis and antibacterial activity of Copper and Nickel saliciliden thiosemicarbazidates. Pharm. Chem. J. 2007, 41, 114–117. [Google Scholar]
- Stavila, V.; Davidovich, R.L.; Gulea, A.; Whitmire, K.H. Bismuth(III) complexes with aminopolycarboxylate and polyaminopolycarboxylate ligands: Chemistry and structure. Coord. Chem. Rev. 2006, 250, 2782–2793. [Google Scholar] [CrossRef]
- Gulea, A.; Poirier, D.; Roy, J.; Tapcov, V. Inhibitors of enzyme 17β-HSD proliferation. Patent of invention MD 3771. BOPI 12, 2008. [Google Scholar]
- Samusi, N.M.; Prisacari, V.I.; Ţapcov, V.I.; Buraciov, S.A.; Gulea, A.P. Synthesis and antimicrobial activity of the complexes of 3d-metals with substituted salicylaldehyde benzoylhydrazones. Pharm. Chem. J. 2004, 38, 373–377. [Google Scholar] [CrossRef]
- Geary, W.J. The use of conductivity measurements in organic solvents for the characterisation of coordination compounds. Coord. Chem. Rev. 1971, 7, 81–122. [Google Scholar] [CrossRef]
- Chumakov, M.Yu.; Janneau, E.; Bejenari, P.N.; Tsapkov, I.V.; Gulea, P.A. Crystal Structure of Cooper Sulfate and Thiocyanate Complexes with 5-Bromo-and 5-Nitrosalicylaldehyde Thiosemicarbazones. Koordinatsionnaya Khimiya 2008, 34, 46–54. [Google Scholar]
- Saydam, S.; Yilmaz, E. Synthesis, characterization and thermal behavior of 4-chloromethyl-2-(2-hydroxybenzilidenehydrazino) thiazole and its complexes with Cr(III), Co(II), Ni(II) and Cu(II). Spectrochim. Acta A 2006, 63, 506–510. [Google Scholar] [CrossRef]
- Larabi, L.; Harek, Y.; Reguig, A.; Mostafa, M.M. Synthesis, structural study and electrochemical properties of copper(II) complexes derived from benzene- and p-toluenesulphonylhydrazones. J. Serb. Chem. Soc. 2003, 68, 85–95. [Google Scholar] [CrossRef]
- Gupta, L.; Chandra, S. Spectroscopy: the study of DNA cleavage by newly synthesized polydentate macrocyclic ligand and its copper(II) complexes. Spectrochim. Acta A 2008, 71, 496–501. [Google Scholar] [CrossRef]
- El-Reash, G.M.A.; Ibrahim, K.M.; Bekheit, M.M. Synthesis, characterization and thermal analysis of new Cu(II) complexes with hydrazide ligands. E-J. Chem. 2010. [Google Scholar] [CrossRef]
- Jouad, E.M.; Riou, A.; Allain, M.; Khan, M.A.; Bouet, G.M. Synthesis, structural and spectral studies of 5-methyl 2-furaldehyde thiosemicarbazone and its Co, Ni, Cu and Cd complexes. Polyhedron 2001, 20, 67–74. [Google Scholar] [CrossRef] [Green Version]
- Chandra, S.; Sangeetika, X. Spectroscopic, redox and biological activities of transition metal complexes with ons donor macrocyclic ligand derived from semicarbazide and thiodiglycolic acid. Spectrochim. Acta A 2004, 60, 147–153. [Google Scholar] [CrossRef]
- Chanrda, S.; Pundir, M. Spectral studies of cobalt(II) complexes of 12-membered macrocyclic ligands having thiosemicarbazone moieties. Spectrochim. Acta A 2007, 68, 833–840. [Google Scholar] [CrossRef]
- Goel, S.; Pandey, O.P.; Sengupta, S.K. Synthesis and physico-chemical studies on neodymium(III) and samarium (III) derivatives with mercapto triazoles. Thermochim. Acta 1988, 133, 359–364. [Google Scholar] [CrossRef]
- Raphae, P.F.; Manoj, E.; Kurup Prathapachandra, M.R. Copper(II) complexes of N(4)-substituted thiosemicarbazones derived from pyridine-2-carbaldehyde: Crystal structure of a binuclear complex. Polyhedron 2007, 26, 818–828. [Google Scholar] [CrossRef]
- Kostas, I.D.; Andreadaki, F.J.; Demertzi, D.K.; Prentjas, C.; Demertzis, M.A. Suzuki-Miyaura cross-coupling reaction of aryl bromides and chlorides with phenylboronic acid under aerobic conditions catalyzed by palladium complexes with thiosemicarbazone ligands. Tetrahedron Lett. 2005, 46, 1967–1970. [Google Scholar] [CrossRef]
- Belicchi-Ferrari, M.; Bisceglie, F.; Cavalieri, C.; Pelosi, G.; Tarasconi, P. Bis(triphenylphosphine)4-fluorobenzaldehyde thiosemicarbazone copper(I): Forcing chelation through oxoanions. Polyhedron 2007, 26, 3774–3782. [Google Scholar] [CrossRef]
- Nakamoto, K. Infrared and Raman Spectra of Inorganic and Coordination Compounds Part B. Applications in Coordination, Organometallic and Bioinorganic Chemisrty, 6th ed.; Wiley-Interscience: Hoboken, NJ, USA, 2008; p. 384. [Google Scholar]
- Drago, R.S. Physical Methods in Chemistry; W.B. Saunders Company: Philadelphia, PA, USA, 1977. [Google Scholar]
- Carlin, R.L. Transition Metal Chemistry, 2nd ed.; Marcel Decker Inc.: New York, NY, USA, 1965. [Google Scholar]
- Pretsch, E.; Clerc, T.; Seibl, J.; Simon, W. Tables of Spectral Data for Structure Determination of Organic Compounds 13C-NMR, 1H-NMR, IR, MS, UV/VIS; Springer-Verlag: Berlin, Gemany, 1989. [Google Scholar]
- Raja, D.S.; Paramaguru, G.; Bhuvanesh, N.S.P.; Reibenspies, J.H.; Renganathan, R.; Natarajan, K. Effect of terminal N- substitution in 2-oxo-1,2-dihydroquinoline-3-carbaldehyde thiosemicarbazone on the mode of ccordination, structure, interaction with protein, radical scavenging and cytotoxic activity of copper(II) complexes. Dalton Trans. 2011, 40, 4548–4559. [Google Scholar] [CrossRef]
- Mohan, M.; Gupta, N.S.; Kumar, A.; Kumar, M. Synthesis, characterization and antitumor activity of iron (II) and iron (III) complexes of 3- and 5- substituted salicylaldehyde benzoyl hydrazones. Inorg. Chim. Acta. 1987, 135, 167–171. [Google Scholar] [CrossRef]
- Mohan, M.; Kumar, A.; Kumar, M.K. Synthesis, characterization and antitumor activity of manganese(II), cobalt(II), nickel(II), copper(II), zinc(II) and platinum complexes of 3- and 5- substituted salicylaldehide benzoylhydrazones. Inorg. Chim. Acta 1987, 136, 65–70. [Google Scholar] [CrossRef]
- Schwarenbach, G.; Flaschka, H. Die Komplexometrische Titration; Ferdinand Euke: Verlag Stuttgart, Germany, 1956. [Google Scholar]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. 2008, A64, 112–122. [Google Scholar]
- Altomare, A.; Burla, M.C.; Camalli, M.; Cascarano, G.L.; Giacovazzo, C.; Guagliardi, A.G.; Moliterni, G.; Polidori, G.; Spagna, R. SIR97: A new tool for crystal structure determination and refinement. J. App. Cryst. 1999, 32, 115–119. [Google Scholar] [CrossRef]
- Betteridge, P.W.; Carruthers, J.R.; Cooper, R.I.; Prout, K.; Watkin, D.J. CRYSTALS version 12: software for guided crystal structure analysis. J. Appl. Cryst. 2003, 36, 1487–1452. [Google Scholar] [CrossRef]
- Sample Availability: Not available.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Pahontu, E.; Fala, V.; Gulea, A.; Poirier, D.; Tapcov, V.; Rosu, T. Synthesis and Characterization of Some New Cu(II), Ni(II) and Zn(II) Complexes with Salicylidene Thiosemicarbazones: Antibacterial, Antifungal and in Vitro Antileukemia Activity. Molecules 2013, 18, 8812-8836. https://doi.org/10.3390/molecules18088812
Pahontu E, Fala V, Gulea A, Poirier D, Tapcov V, Rosu T. Synthesis and Characterization of Some New Cu(II), Ni(II) and Zn(II) Complexes with Salicylidene Thiosemicarbazones: Antibacterial, Antifungal and in Vitro Antileukemia Activity. Molecules. 2013; 18(8):8812-8836. https://doi.org/10.3390/molecules18088812
Chicago/Turabian StylePahontu, Elena, Valeriu Fala, Aurelian Gulea, Donald Poirier, Victor Tapcov, and Tudor Rosu. 2013. "Synthesis and Characterization of Some New Cu(II), Ni(II) and Zn(II) Complexes with Salicylidene Thiosemicarbazones: Antibacterial, Antifungal and in Vitro Antileukemia Activity" Molecules 18, no. 8: 8812-8836. https://doi.org/10.3390/molecules18088812
APA StylePahontu, E., Fala, V., Gulea, A., Poirier, D., Tapcov, V., & Rosu, T. (2013). Synthesis and Characterization of Some New Cu(II), Ni(II) and Zn(II) Complexes with Salicylidene Thiosemicarbazones: Antibacterial, Antifungal and in Vitro Antileukemia Activity. Molecules, 18(8), 8812-8836. https://doi.org/10.3390/molecules18088812













