A Modular Approach to Triazole-Containing Chemical Inducers of Dimerisation for Yeast Three-Hybrid Screening
Abstract
:1. Introduction
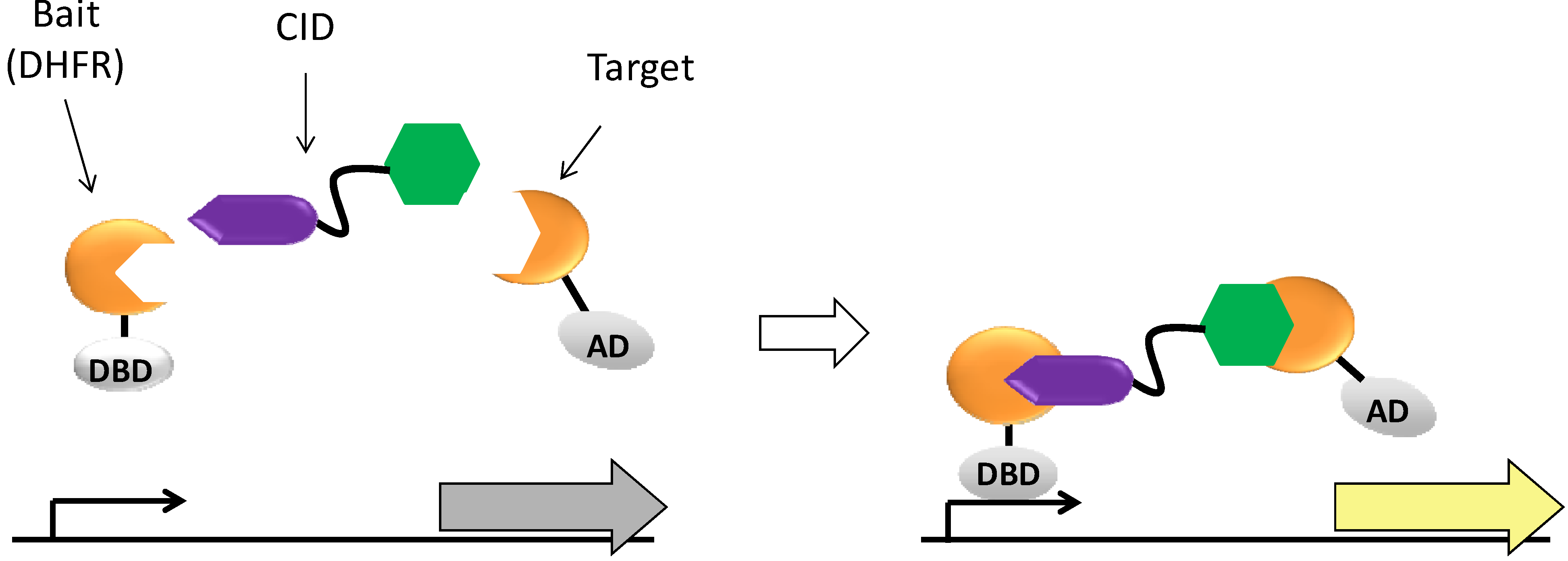
2. Results and Discussion
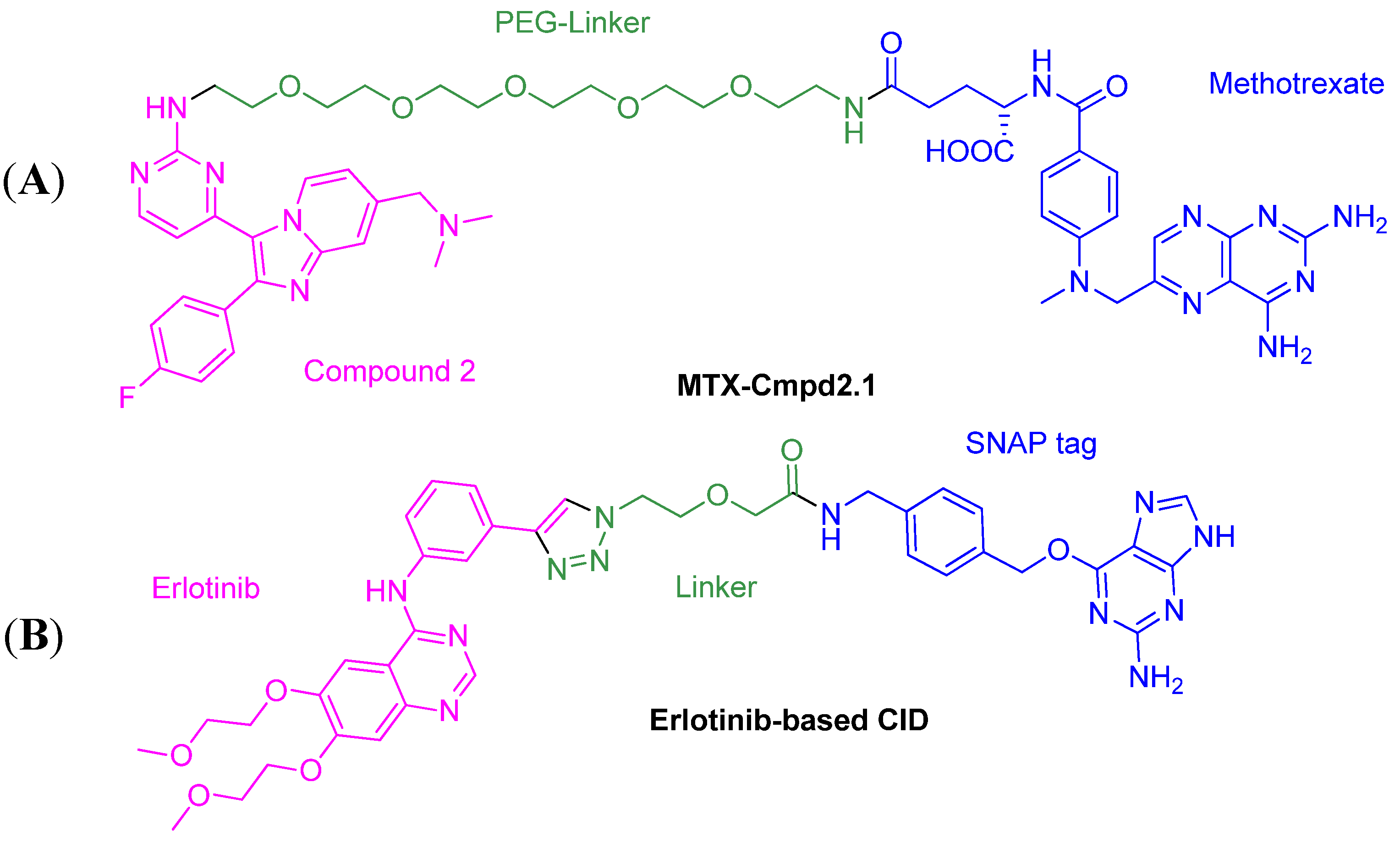
2.1. Comparison of MTX-Cmpd2.1 (PEG Linker) and MTX-Cmpd2.2 (Triazole Linker)
2.1.1. Synthesis of the Compound 2-based Alkynes 3
2.1.2. Synthesis of the tBu-MTX-Azides 4
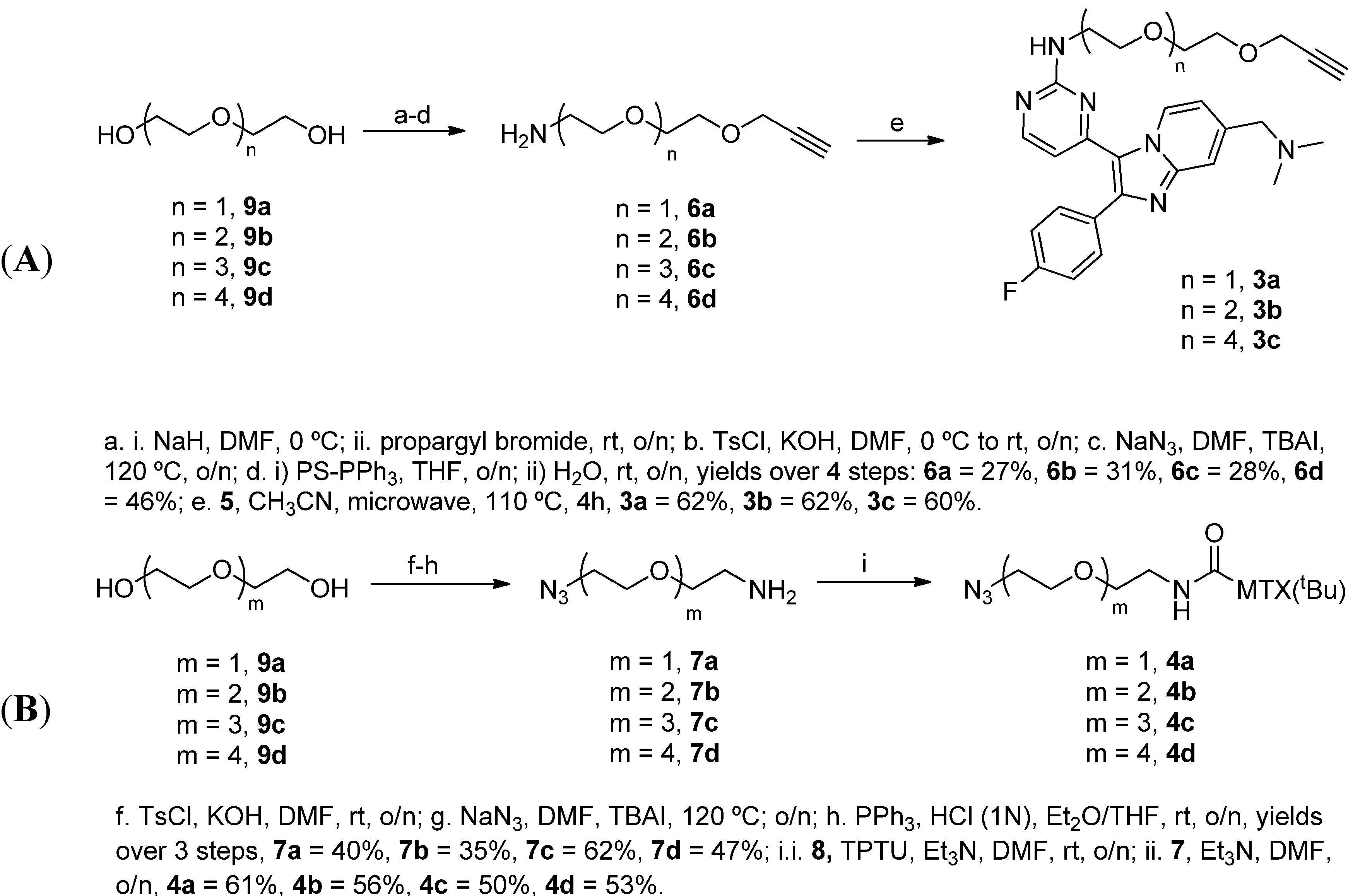
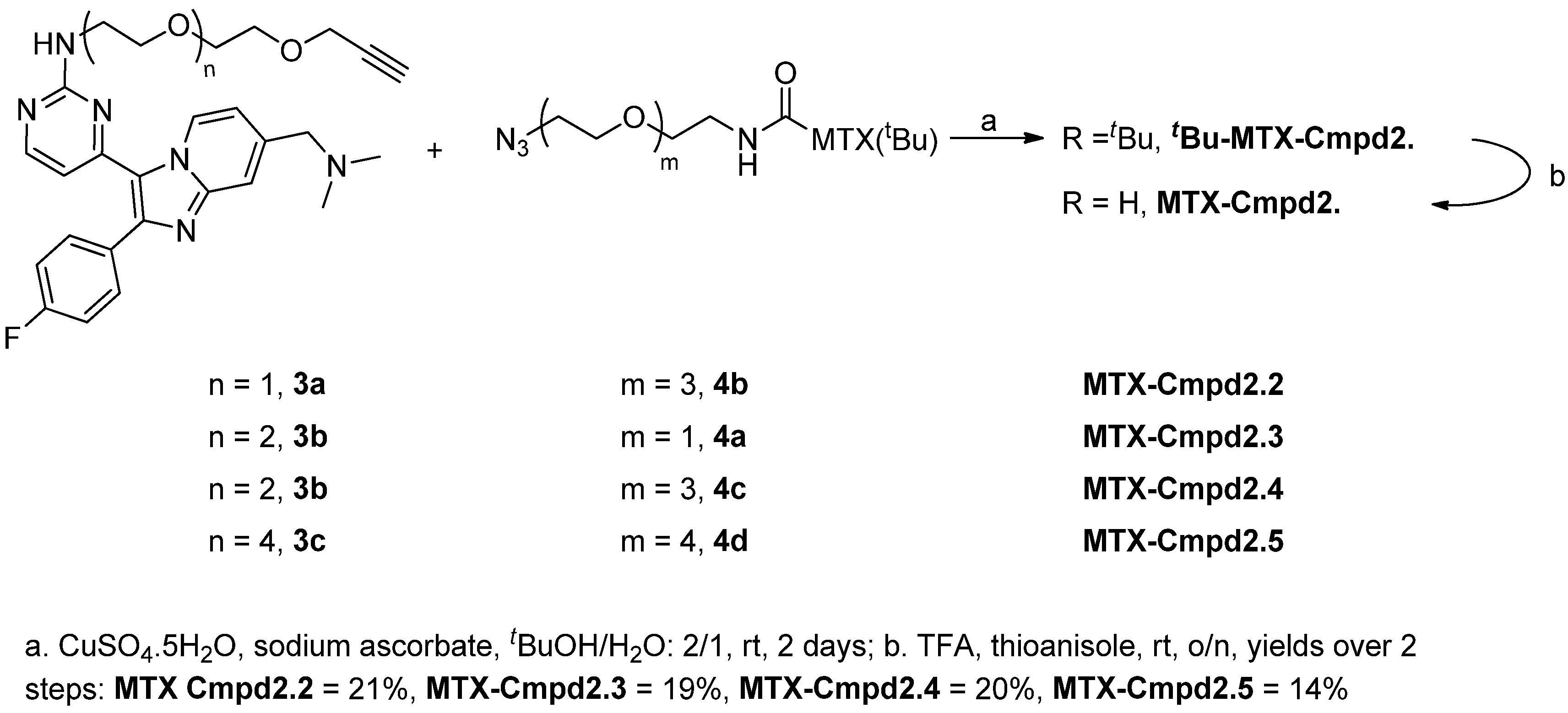
2.1.3. Assembly of CID MTX-Cmpd2.2
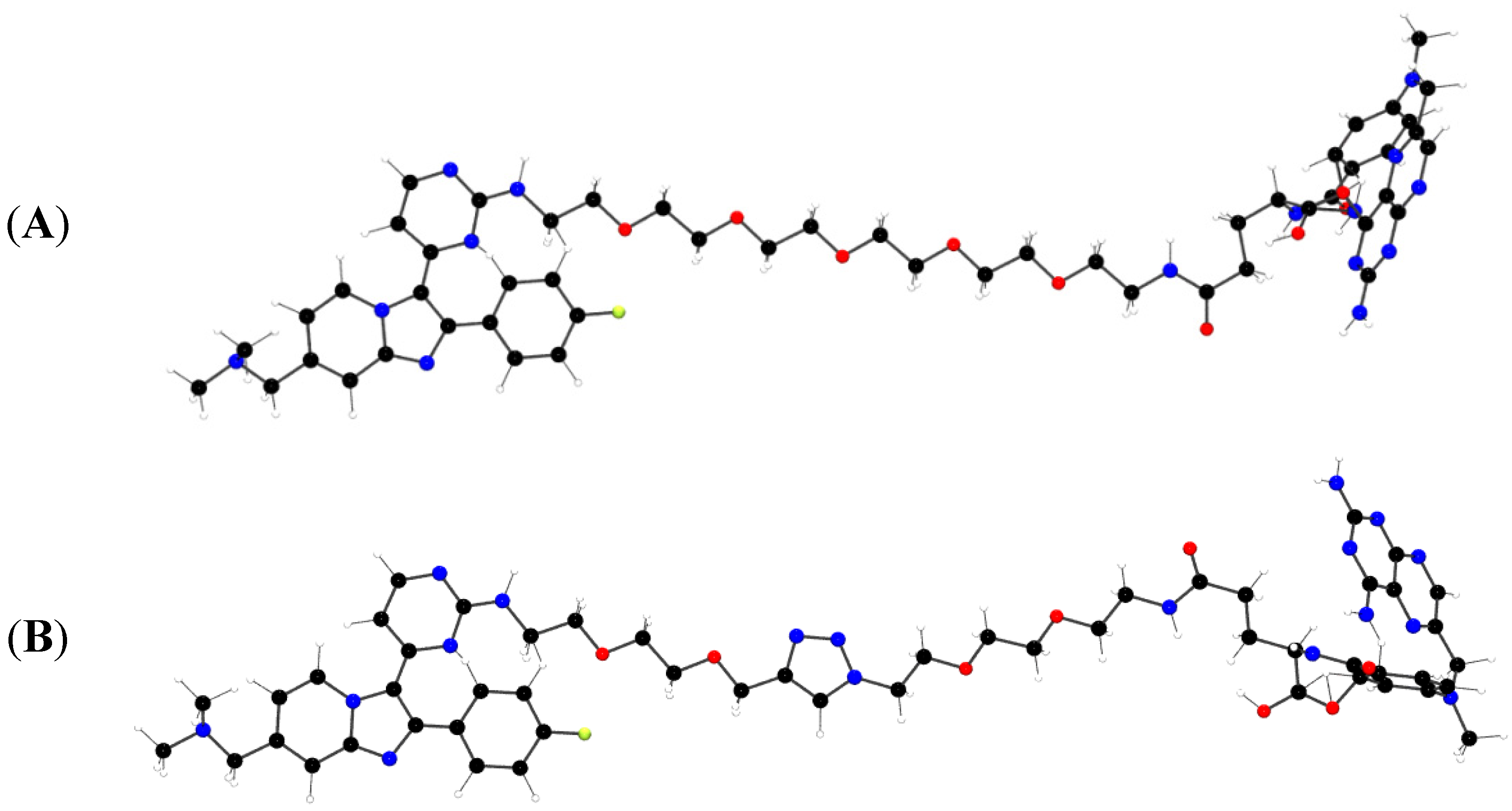
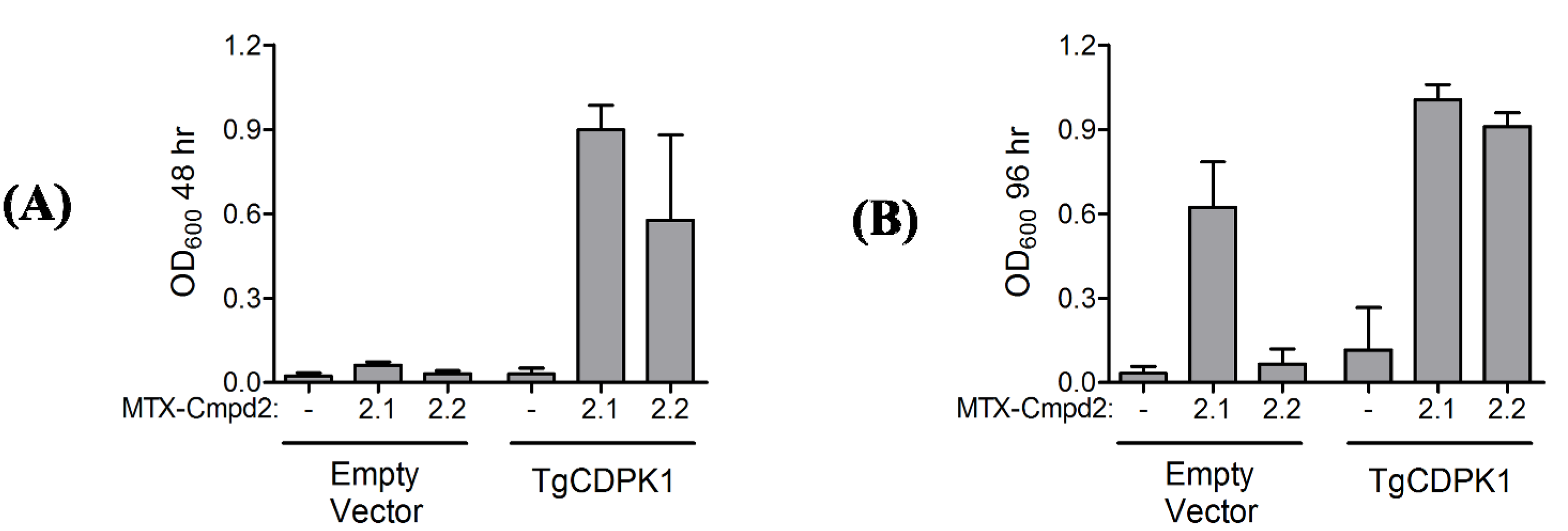
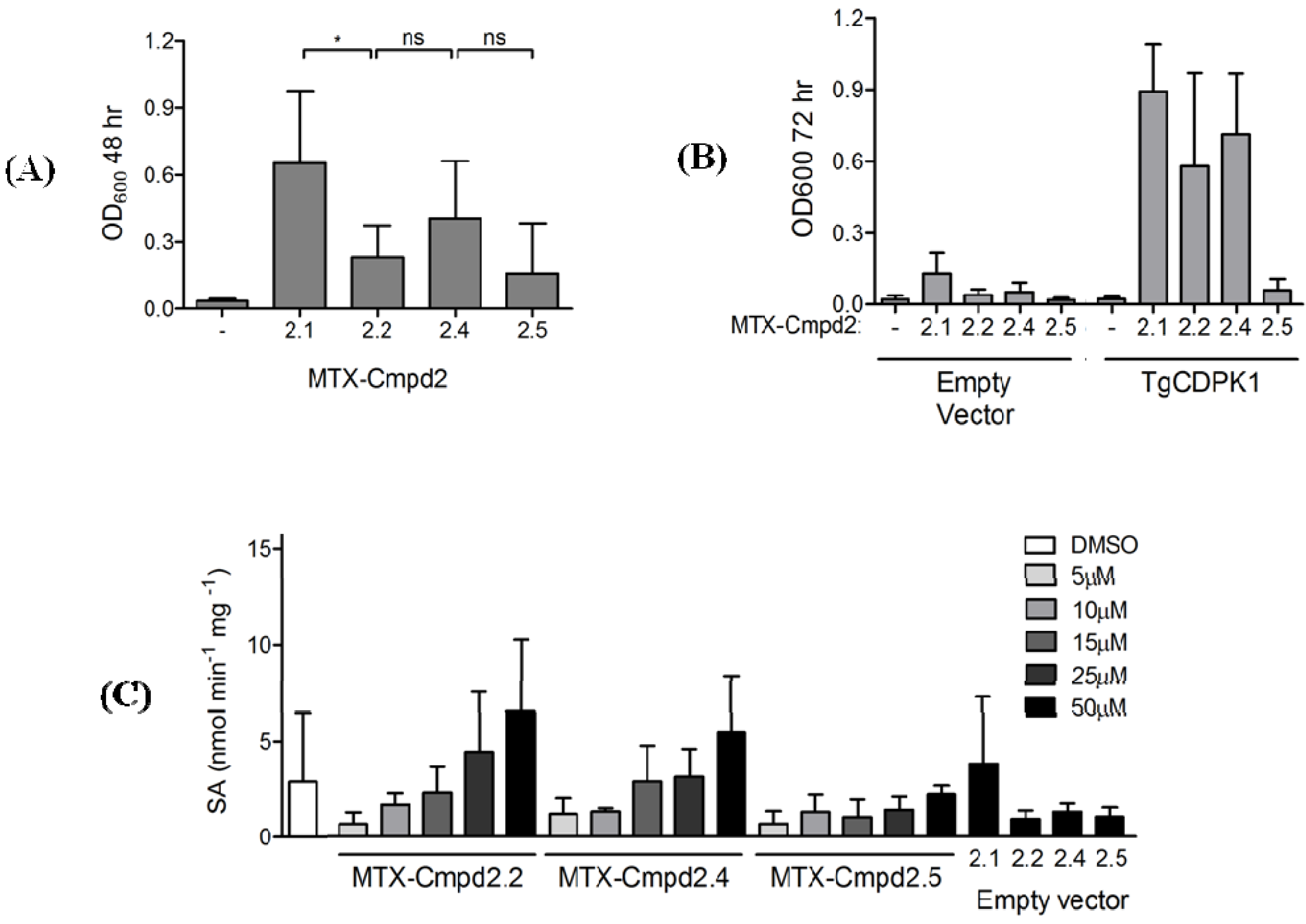
2.1.4. Y3H results with CIDs MTX-Cmpd2.1 and MTX-Cmpd2.2
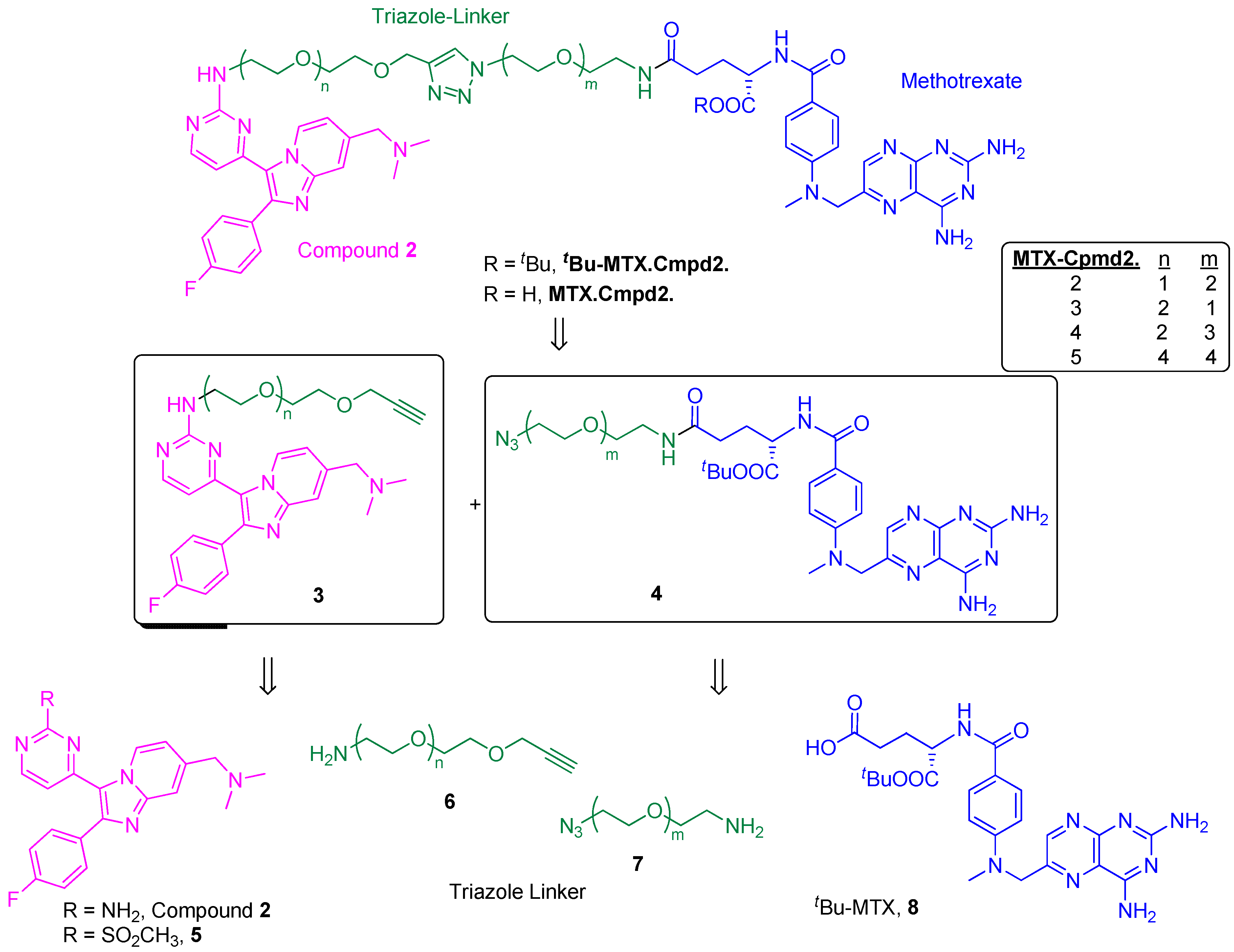
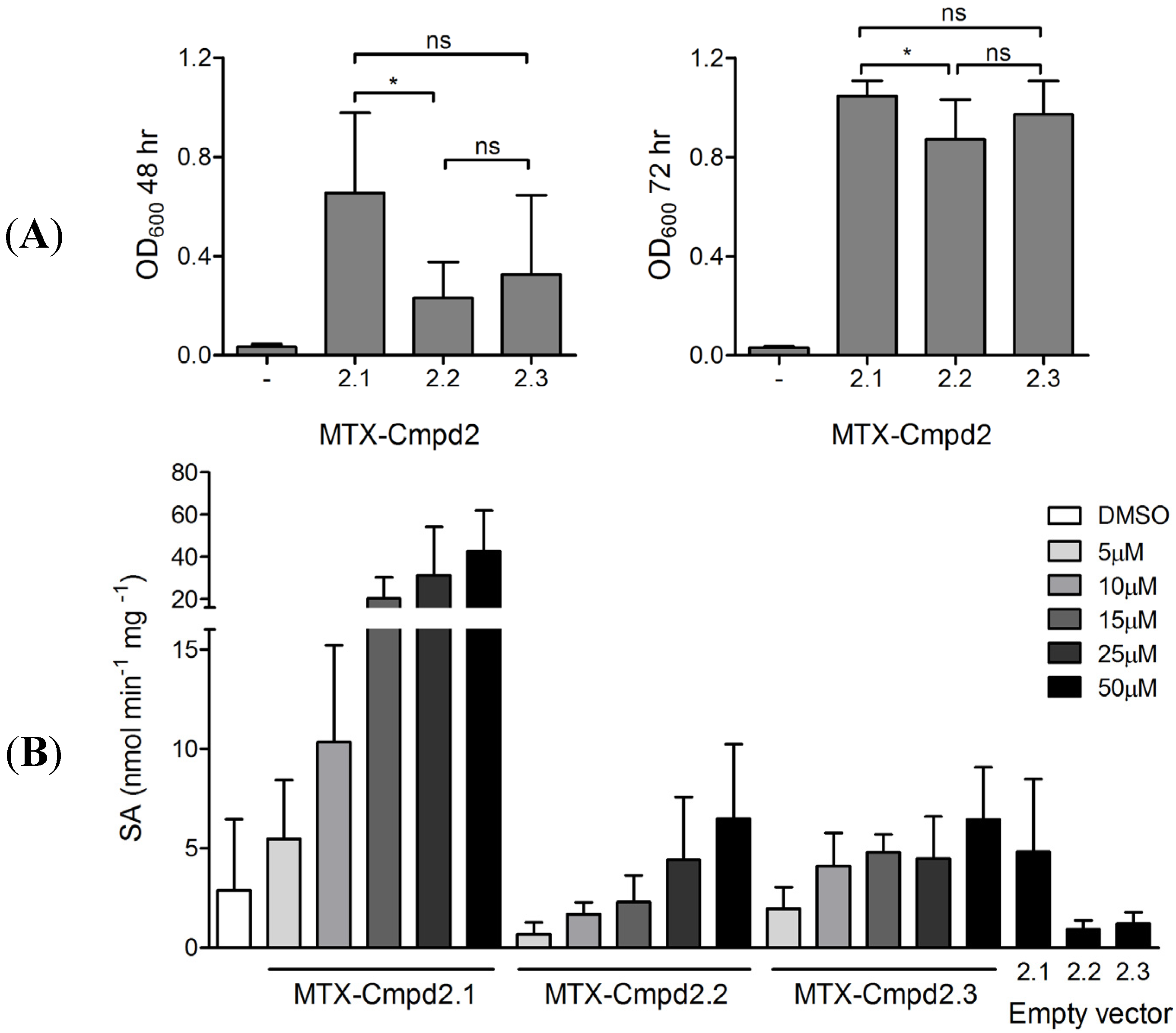
2.2. Synthesis and Analysis of Additional MTX-Cmpd2 CIDs
3. Experimental
3.1. General
3.2. Synthetic Procedures
3.2.1. General Procedure A: Synthesis of Compound 2-based alkyne 3
3.2.2. General Procedure B: Synthesis of tBu-MTX-azide Components 4
3.2.3. General Procedure C: Coupling of 3 and 4
3.2.4. General Procedure D: Boc Deprotection of tBu-MTX-Cmpd2
3.3. Reporter Activation Assays
4. Conclusions
Supplementary Materials
Acknowledgments
Conflicts of Interest
References
- Althoff, E.A.; Cornish, V.W. A Bacterial Small-Molecule Three-Hybrid System. Angew. Chem. Int. Ed. 2002, 41, 2327–2330. [Google Scholar] [CrossRef]
- Baker, K.; Sengupta, D.; Salazar-Jimenez, G.; Cornish, V.W. An optimized dexamethasone-methotrexate yeast 3-hybrid system for high-throughput screening of small molecule-protein interactions. Anal. Biochem. 2003, 315, 134–137. [Google Scholar] [CrossRef]
- Becker, F.; Murthi, K.; Smith, C.; Come, J.; Costa-Roldán, N.; Kaufmann, C.; Hanke, U.; Degenhart, C.; Baumann, S.; Wallner, W.; et al. A Three-Hybrid Approach to Scanning the Proteome for Targets of Small Molecule Kinase Inhibitors. Chem. Biol. 2004, 11, 211–223. [Google Scholar]
- Caligiuri, M.; Becker, F.; Murthi, K.; Kaplan, F.; Dedier, S.; Kaufmann, C.; Machl, A.; Zybarth, G.; Richard, J.; Bockovich, N.; Kluge, A.; Kley, N. A Proteome-Wide CDK/CRK-Specific Kinase Inhibitor Promotes Tumor Cell Death in the Absence of Cell Cycle Progression. Chem. Biol. 2005, 12, 1103–1115. [Google Scholar] [CrossRef]
- Caligiuri, M.; Molz, L.; Liu, Q.; Kaplan, F.; Xu, J.P.; Majeti, J.Z.; Ramos-Kelsey, R.; Murthi, K.; Lievens, S.; Tavernier, J.; Kley, N. MASPIT: Three-Hybrid Trap for Quantitative Proteome Fingerprinting of Small Molecule-Protein Interactions in Mammalian Cells. Chem. Biol. 2006, 13, 711–722. [Google Scholar] [CrossRef]
- Chidley, C.; Haruki, H.; Pedersen, M.G.N.; Muller, E.; Johnsson, K. A yeast-based screen reveals that sulfasalazine inhibits tetrahydrobiopterin biosynthesis. Nat. Chem. Biol. 2011, 7, 375–383. [Google Scholar] [CrossRef]
- Corson, T.W.; Aberle, N.; Crews, C.M. Design and Applications of Bifunctional Small Molecules: Why Two Heads Are Better Than One. ACS Chem. Biol. 2008, 3, 677–692. [Google Scholar] [CrossRef]
- Cottier, S.; Mönig, T.; Wang, Z.; Svoboda, J.; Boland, W.; Kaiser, M.; Kombrink, E. The Yeast Three-Hybrid System as an Experimental Platform to Identify Proteins Interacting with Small Signaling Molecules in Plant Cells: Potential and Limitations. Front. Plant Sci. 2011, 2, 1–12. [Google Scholar]
- Donald, R.G.K.; Zhong, T.; Wiersma, H.; Nare, B.; Yao, D.; Lee, A.; Allocco, J.; Liberator, P.A. Anticoccidial kinase inhibitors: Identification of protein kinase targets secondary to cGMP-dependent protein kinase. Mol. Biochem. Parasitol. 2006, 149, 86–98. [Google Scholar] [CrossRef]
- Jaeger, S.; Eriani, G.; Martin, F. Results and prospects of the yeast three-hybrid system. FEBS Lett. 2004, 556, 7–12. [Google Scholar] [CrossRef]
- Kley, N. Chemical Dimerizers and Three-Hybrid Systems: Scanning the Proteome for Targets of Organic Small Molecules. Chem. Biol. 2004, 11, 599–608. [Google Scholar]
- Lefurgy, S.; Cornish, V. Finding Cinderella after the Ball: A Three-Hybrid Approach to Drug Target Identification. Chem. Biol. 2004, 11, 151–153. [Google Scholar]
- Licitra, E.J.; Liu, J.O. A three-hybrid system for detecting small ligand-protein receptor interactions. Proc. Natl. Acad. Sci. 1996, 93, 12817–12821. [Google Scholar] [CrossRef]
- Maniataki, E.; Martinez de Alba, A.; Sägesser, R.; Tabler, M.; Tsagris, M. Viroid RNA systemic spread may depend on the interaction of a 71-nucleotide bulged hairpin with the host protein VirP1. RNA 2003, 9, 346–354. [Google Scholar] [CrossRef]
- Schneider, K.; Wang, Z.M.; Kaiser, E.; Kombrink, E. On the molecular mechanism of hormone action: hunting the jasmonate target(s). Curr. Topics Phytochem. 2008, 9, 1–16. [Google Scholar]
- Vollmeister, E.; Haag, C.; Zarnack, K.; Baumann, S.; König, J.; Mannhaupt, G.; Feldbrügge, M. Tandem KH domains of Khd4 recognize AUACCC and are essential for regulation of morphology as well as pathogenicity in Ustilago maydis. RNA 2009, 15, 2206–2218. [Google Scholar] [CrossRef]
- Wurster, S.E.; Maher, L.J. Selections that optimize RNA display in the yeast three-hybrid system. RNA 2010, 16, 253–258. [Google Scholar] [CrossRef]
- Shepard, A.R.; Conrow, R.E.; Pang, I.-H.; Jacobson, N.; Rezwan, M.; Rutschmann, K.; Auerbach, D.; SriRamaratnam, R.; Cornish, V.W. Identification of PDE6D as a Molecular Target of Anecortave Acetate via a Methotrexate-Anchored Yeast Three-Hybrid Screen. ACS Chem. Biol. 2013, 8, 549–558. [Google Scholar] [CrossRef]
- Abida, W.M.; Carter, B.T.; Althoff, E.A.; Lin, H.; Cornish, V.W. Receptor-Dependence of the Transcription Read-Out in a Small-Molecule Three-Hybrid System. ChemBioChem 2002, 3, 887–895. [Google Scholar] [CrossRef]
- Henthorn, D.C.; Jaxa-Chamiec, A.A.; Meldrum, E. A GAL4-based yeast three-hybrid system for the identification of small molecule–target protein interactions. Biochem. Pharmacol. 2002, 63, 1619–1628. [Google Scholar] [CrossRef]
- Lin, H.; Abida, W.M.; Sauer, R.T.; Cornish, V.W. Dexamethasone−Methotrexate: An Efficient Chemical Inducer of Protein Dimerization In Vivo. J. Am. Chem. Soc. 2000, 122, 4247–4248. [Google Scholar] [CrossRef]
- Suwwan de Felipe, K.; Carter, B.T.; Althoff, E.A.; Cornish, V.W. Correlation between Ligand-Receptor Affinity and the Transcription Readout in a Yeast Three-Hybrid System. Biochemistry 2004, 43, 10353–10363. [Google Scholar] [CrossRef]
- Odell, A.V.; Tran, F.; Poupart, S.; Pathak, R.; Westwood, N.J.; Ward, G.E. Yeast three-hybrid screen identifies TgBRADIN as a novel negative regulator of Toxoplasma gondii bradyzoite differentiation. PLoS One 2013. submitted. [Google Scholar]
- Walton, J.G. A.; Patterson, S.; Liu, G.; Haraldsen, J.D.; Hollick, J.J.; Slawin, A.M.Z.; Ward, G.E.; Westwood, N.J. Synthesis and biological evaluation of functionalised tetrahydro-[small beta]-carboline analogues as inhibitors of Toxoplasma gondii invasion. Org. Biomol. Chem. 2009, 7, 3049–3060. [Google Scholar] [CrossRef]
- Amara, J.F.; Clackson, T.; Rivera, V.M.; Guo, T.; Keenan, T.; Natesan, S.; Pollock, R.; Yang, W.; Courage, N.L.; Holt, D.A.; Gilman, M. A versatile synthetic dimerizer for the regulation of protein–protein interactions. Proc. Natl. Acad. Sci. 1997, 94, 10618–10623. [Google Scholar] [CrossRef]
- Spencer, D.M.; Wandless, T.J; Schreiber, S.L.; Crabtree, G.R. Controlling signal transduction with synthetic ligands. Science 1993, 262, 1019–1024. [Google Scholar]
- Lu, G.; Lam, S.; Burgess, K. An iterative route to “decorated” ethylene glycol-based linkers. Chem. Commun. 2006, 2006, 1652–1654. [Google Scholar]
- Biftu, T.; Feng, D.; Fisher, M.; Liang, G.-B.; Qian, X.; Scribner, A.; Dennis, R.; Lee, S.; Liberator, P.A.; Brown, C.; et al. Synthesis and SAR studies of very potent imidazopyridine antiprotozoal agents. Bioorg. Med. Chem. Lett. 2006, 16, 2479–2483. [Google Scholar] [CrossRef]
- Bonger, K.M.; van den Berg, R.J.; Heitman, L.H.; Ijzerman, A.P.; Oosterom, J.; Timmers, C.M.; Overkleeft, H.S.; van der Marel, G.A. Synthesis and evaluation of homo-bivalent GnRHR ligands. Bioorg. Med. Chem. 2007, 15, 4841–4856. [Google Scholar] [CrossRef]
- Gervay-Hague, J.; Du, W.; Denardo, S.; Natarajan, A. Construction of a multivalent SCFV through alkyne-azide 1,3-dipolar cycloaddition. WO 2007/112362 A2, 4 October 2007. [Google Scholar]
- Klein, E.; DeBonis, S.; Thiede, B.; Skoufias, D.A.; Kozielski, F.; Lebeau, L. New chemical tools for investigating human mitotic kinesin Eg5. Bioorg. Med. Chem. 2007, 15, 6474–6488. [Google Scholar] [CrossRef]
- Kolb, H.C.; Finn, M.G.; Sharpless, K.B. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew. Chem. Int. Ed. 2001, 40, 2004–2021. [Google Scholar] [CrossRef]
- Kolb, H.C.; Sharpless, K.B. The growing impact of click chemistry on drug discovery. Drug Discov. Today 2003, 8, 1128–1137. [Google Scholar] [CrossRef]
- Rostovtsev, V.V.; Green, L.G.; Fokin, V.V.; Sharpless, K.B. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective “Ligation” of Azides and Terminal Alkynes. Angew. Chem. Int. Ed. 2002, 41, 2596–2599. [Google Scholar]
- Thirumurugan, P.; Matosiuk, D.; Jozwiak, K. Click Chemistry for Drug Development and Diverse Chemical–Biology Applications. Chem. Rev. 2013, 113, 4905–4979. [Google Scholar] [CrossRef]
- Robinson, C.R.; Sauer, R.T. Optimizing the stability of single-chain proteins by linker length and composition mutagenesis. Proc. Natl. Acad. Sci. 1998, 95, 5929–5934. [Google Scholar] [CrossRef]
- George, R.A.; Heringa, J. An analysis of protein domain linkers: their classification and role in protein folding. Protein Eng. 2002, 15, 871–879. [Google Scholar] [CrossRef]
- Nusinow, D.A.; Helfer, A.; Hamilton, E.E.; King, J.J.; Imaizumi, T.; Schultz, T.F.; Farre, E.M.; Kay, S.A. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 2011, 475, 398–402. [Google Scholar] [CrossRef]
- Rietz, S.; Stamm, A.; Malonek, S.; Wagner, S.; Becker, D.; Medina-Escobar, N.; Corina Vlot, A.; Feys, B.J.; Niefind, K.; Parker, J.E. Different roles of Enhanced Disease Susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 2011, 191, 107–119. [Google Scholar] [CrossRef]
- Sheerin, D.J.; Buchanan, J.; Kirk, C.; Harvey, D.; Sun, X.; Spagnuolo, J.; Li, S.; Liu, T.; Woods, V.A.; Foster, T.; Jones, W.T.; Rakonjac, J. Inter- and intra-molecular interactions of Arabidopsis thaliana DELLA protein RGL1. Biochem. J. 2011, 435, 629–639. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds 3a–c and 4a–d are available from the authors.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Tran, F.; Odell, A.V.; Ward, G.E.; Westwood, N.J. A Modular Approach to Triazole-Containing Chemical Inducers of Dimerisation for Yeast Three-Hybrid Screening. Molecules 2013, 18, 11639-11657. https://doi.org/10.3390/molecules180911639
Tran F, Odell AV, Ward GE, Westwood NJ. A Modular Approach to Triazole-Containing Chemical Inducers of Dimerisation for Yeast Three-Hybrid Screening. Molecules. 2013; 18(9):11639-11657. https://doi.org/10.3390/molecules180911639
Chicago/Turabian StyleTran, Fanny, Anahi V. Odell, Gary E. Ward, and Nicholas J. Westwood. 2013. "A Modular Approach to Triazole-Containing Chemical Inducers of Dimerisation for Yeast Three-Hybrid Screening" Molecules 18, no. 9: 11639-11657. https://doi.org/10.3390/molecules180911639




