Applications of Copper-Catalyzed Click Chemistry in Activity-Based Protein Profiling
Abstract
:1. Introduction


2. The Development of CuAAC and Early Applications to ABPP
3. Alkyne-Tagged ABPs for the Serine Hydrolase Family

4. CuAAC-Compatible Covalent Probes for Other Enzyme Families
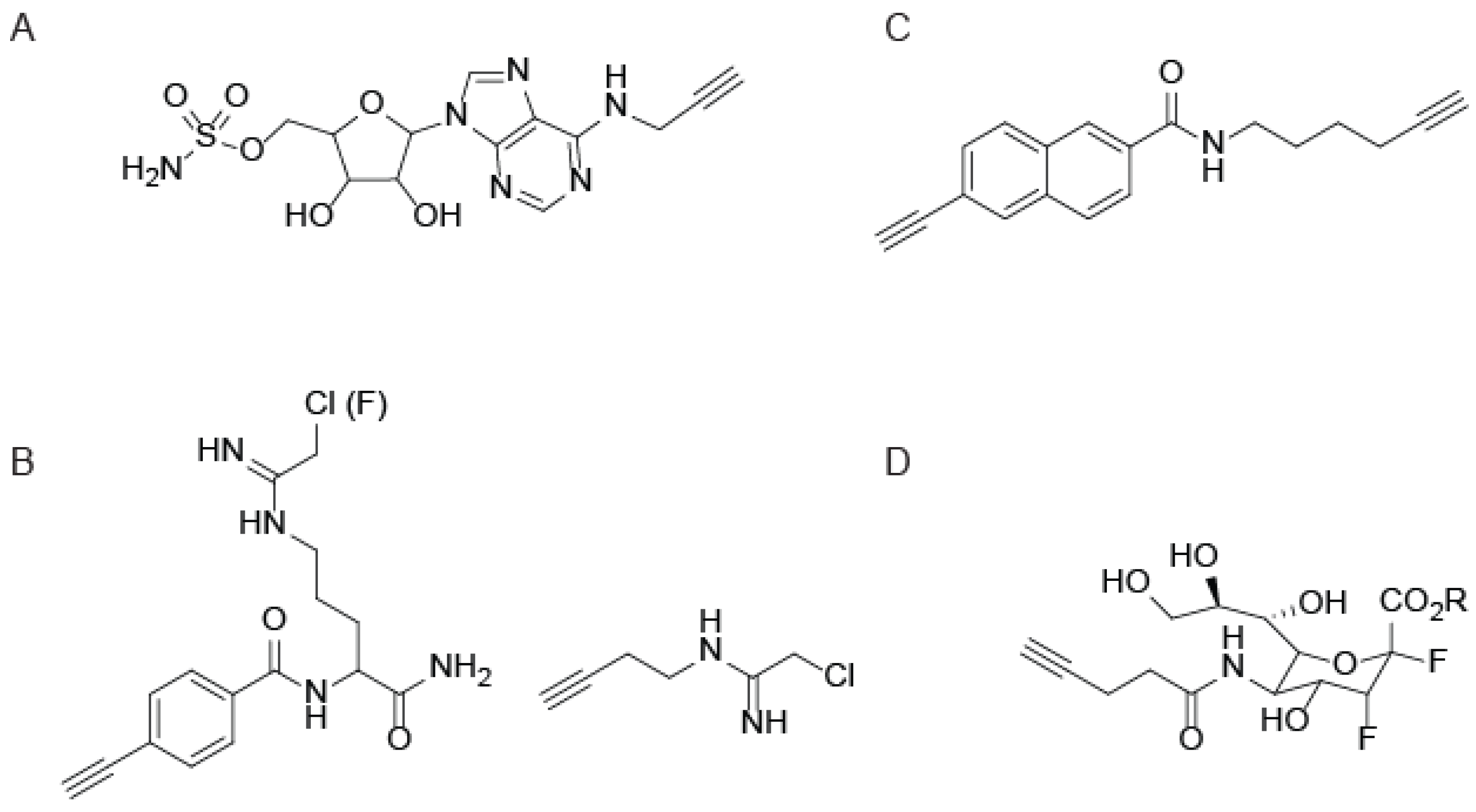
5. Photo-crosslinking ABPs that Utilize CuAAC
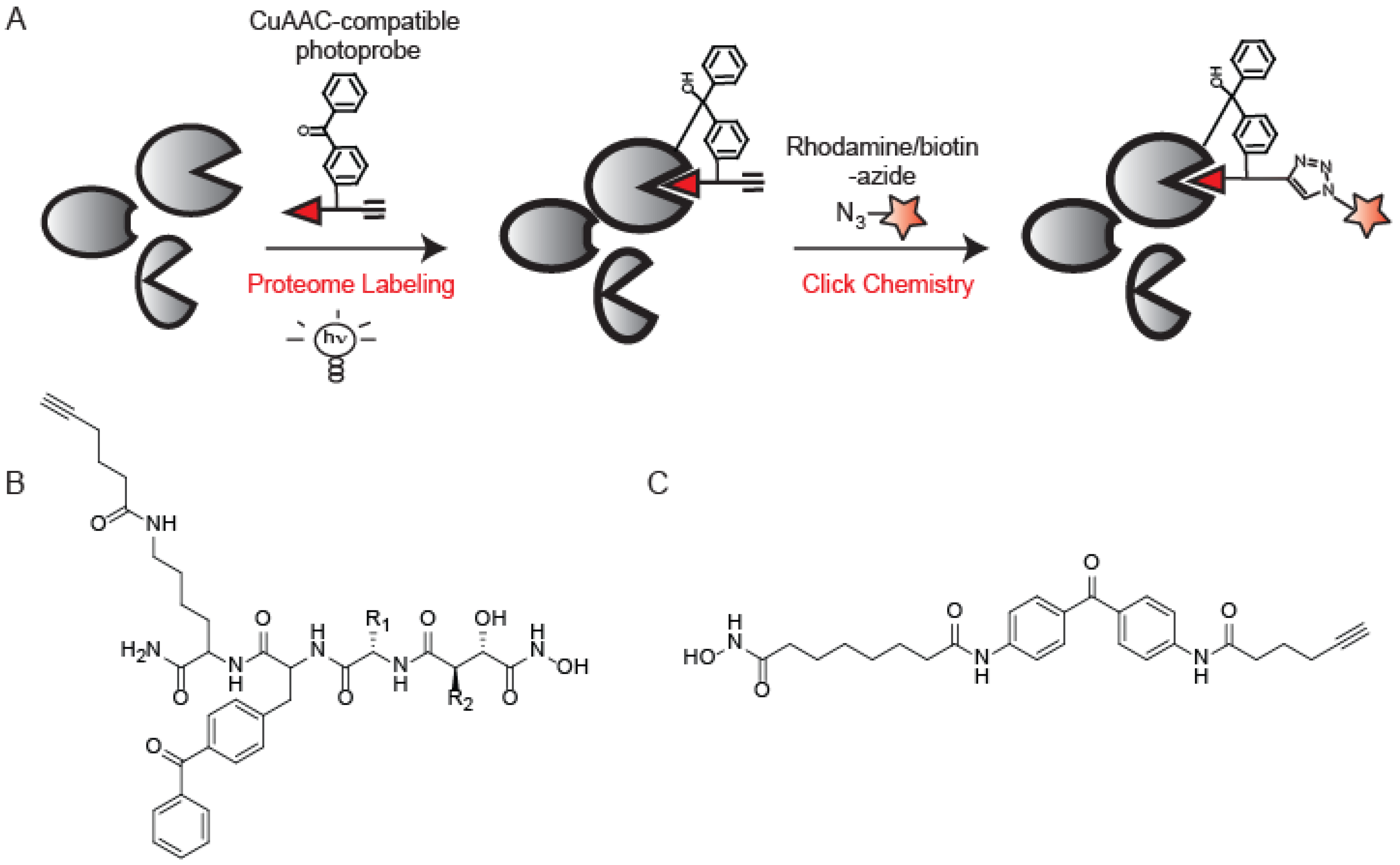
6. Applications of CuAAC in Mass Spectrometry-Based ABPP

7. Conclusions
Conflicts of Interest
References
- Zhang, Y.; Fonslow, B.R.; Shan, B.; Baek, M.C.; Yates, J.R., 3rd. Protein analysis by shotgun/bottom-up proteomics. Chem. Rev. 2013, 113, 2343–2394. [Google Scholar] [CrossRef]
- Kobe, B.; Kemp, B.E. Active site-directed protein regulation. Nature 1999, 402, 373–376. [Google Scholar] [CrossRef]
- Adam, G.C.; Sorensen, E.J.; Cravatt, B.F. Chemical strategies for functional proteomics. Mol. Cell. Proteomics 2002, 1, 781–790. [Google Scholar] [CrossRef]
- Cravatt, B.F.; Sorensen, E.J. Chemical strategies for the global analysis of protein function. Curr. Opin. Chem. Biol. 2000, 4, 663–668. [Google Scholar] [CrossRef]
- Sadaghiani, A.M.; Verhelst, S.H.; Bogyo, M. Tagging and detection strategies for activity-based proteomics. Curr. Opin. Chem. Biol. 2007, 11, 20–28. [Google Scholar] [CrossRef]
- Fonovic, M.; Bogyo, M. Activity-based probes as a tool for functional proteomic analysis of proteases. Expert Rev. Proteomics 2008, 5, 721–730. [Google Scholar] [CrossRef]
- Evans, M.J.; Cravatt, B.F. Mechanism-based profiling of enzyme families. Chem. Rev. 2006, 106, 3279–3301. [Google Scholar] [CrossRef]
- Cravatt, B.F.; Wright, A.T.; Kozarich, J.W. Activity-based protein profiling: From enzyme chemistry. Annu. Rev. Biochem. 2008, 77, 383–414. [Google Scholar] [CrossRef]
- Speers, A.E.; Cravatt, B.F. Chemical strategies for activity-based proteomics. Chembiochem 2004, 5, 41–47. [Google Scholar] [CrossRef]
- Speers, A.E.; Adam, G.C.; Cravatt, B.F. Activity-based protein profiling in vivo using a copper(i)-catalyzed azide-alkyne [3 + 2] cycloaddition. J. Am. Chem. Soc. 2003, 125, 4686–4687. [Google Scholar] [CrossRef]
- Speers, A.E.; Cravatt, B.F. Profiling enzyme activities in vivo using click chemistry methods. Chem. Biol. 2004, 11, 535–546. [Google Scholar] [CrossRef]
- Speers, A.E.; Cravatt, B.F. Activity-based protein profiling (ABPP) and click chemistry (CC)-ABPP by MudPIT mass spectrometry. Curr. Protoc. Chem. Biol. 2009, 1, 29–41. [Google Scholar]
- Willems, L.I.; van der Linden, W.A.; Li, N.; Li, K.Y.; Liu, N.; Hoogendoorn, S.; van der Marel, G.A.; Florea, B.I.; Overkleeft, H.S. Bioorthogonal chemistry: Applications in activity-based protein profiling. Acc. Chem. Res. 2011, 44, 718–729. [Google Scholar] [CrossRef]
- Ramil, C.P.; Lin, Q. Bioorthogonal chemistry: Strategies and recent developments. Chem. Commun. 2013, 49, 11007–11022. [Google Scholar] [CrossRef]
- Saxon, E.; Armstrong, J.I.; Bertozzi, C.R. A “traceless” Staudinger ligation for the chemoselective synthesis of amide bonds. Org. Lett. 2000, 2, 2141–2143. [Google Scholar] [CrossRef]
- Saxon, E.; Bertozzi, C.R. Cell surface engineering by a modified Staudinger reaction. Science 2000, 287, 2007–1010. [Google Scholar] [CrossRef]
- Devaraj, N.K.; Weissleder, R. Biomedical applications of tetrazine cycloadditions. Acc. Chem. Res. 2011, 44, 816–827. [Google Scholar] [CrossRef]
- Devaraj, N.K.; Upadhyay, R.; Haun, J.B.; Hilderbrand, S.A.; Weissleder, R. Fast and sensitive pretargeted labeling of cancer cells through a tetrazine/trans-cyclooctene cycloaddition. Angew. Chem. Int. Ed. Engl. 2009, 48, 7013–7016. [Google Scholar] [CrossRef]
- Devaraj, N.K.; Weissleder, R.; Hilderbrand, S.A. Tetrazine-based cycloadditions: Application to pretargeted live cell imaging. Bioconjug. Chem. 2008, 19, 2297–2299. [Google Scholar] [CrossRef]
- Tornoe, C.W.; Christensen, C.; Meldal, M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(i)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 2002, 67, 3057–3064. [Google Scholar] [CrossRef]
- Wang, Q.; Chan, T.R.; Hilgraf, R.; Fokin, V.V.; Sharpless, K.B.; Finn, M.G. Bioconjugation by copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition. J. Am. Chem. Soc. 2003, 125, 3192–3193. [Google Scholar] [CrossRef]
- Chang, P.V.; Prescher, J.A.; Sletten, E.M.; Baskin, J.M.; Miller, I.A.; Agard, N.J.; Lo, A.; Bertozzi, C.R. Copper-free click chemistry in living animals. Proc. Natl. Acad. Sci. USA 2010, 107, 1821–1826. [Google Scholar] [CrossRef]
- Baskin, J.M.; Prescher, J.A.; Laughlin, S.T.; Agard, N.J.; Chang, P.V.; Miller, I.A.; Lo, A.; Codelli, J.A.; Bertozzi, C.R. Copper-free click chemistry for dynamic in vivo imaging. Proc. Natl. Acad. Sci. USA 2007, 104, 16793–16797. [Google Scholar] [CrossRef]
- Agard, N.J.; Prescher, J.A.; Bertozzi, C.R. A strain-promoted [3 + 2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 2004, 126, 15046–15047. [Google Scholar] [CrossRef]
- Hong, V.; Steinmetz, N.F.; Manchester, M.; Finn, M.G. Labeling live cells by copper-catalyzed alkyne–azide click chemistry. Bioconjug. Chem. 2010, 21, 1912–1916. [Google Scholar] [CrossRef]
- Huisgen, R. 1,3,-Dipolar Cycloaddition Chemistry; Wiley: New York, NY, USA, 1984; pp. 1–176. [Google Scholar]
- Rostovtsev, V.V.; Green, L.G.; Fokin, V.V.; Sharpless, K.B. A stepwise huisgen cycloaddition process: Copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. Engl. 2002, 41, 2596–2599. [Google Scholar] [CrossRef]
- Long, J.Z.; Cravatt, B.F. The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem. Rev. 2011, 111, 6022–6063. [Google Scholar] [CrossRef]
- Dodson, G.; Wlodawer, A. Catalytic triads and their relatives. Trends Biochem. Sci. 1998, 23, 347–352. [Google Scholar] [CrossRef]
- Kidd, D.; Liu, Y.; Cravatt, B.F. Profiling serine hydrolase activities in complex proteomes. Biochemistry 2001, 40, 4005–4015. [Google Scholar] [CrossRef]
- Liu, Y.; Patricelli, M.P.; Cravatt, B.F. Activity-based protein profiling: The serine hydrolases. Proc. Natl. Acad. Sci. USA 1999, 96, 14694–14699. [Google Scholar] [CrossRef]
- Simon, G.M.; Cravatt, B.F. Activity-based proteomics of enzyme superfamilies: Serine hydrolases as a case study. J. Biol. Chem. 2010, 285, 11051–11055. [Google Scholar] [CrossRef]
- Gillet, L.C.; Namoto, K.; Ruchti, A.; Hoving, S.; Boesch, D.; Inverardi, B.; Mueller, D.; Coulot, M.; Schindler, P.; Schweigler, P.; et al. In-cell selectivity profiling of serine protease inhibitors by activity-based proteomics. Mol. Cell. Proteomics 2008, 7, 1241–1253. [Google Scholar] [CrossRef]
- Tully, S.E.; Cravatt, B.F. Activity-based probes that target functional subclasses of phospholipases in proteomes. J. Am. Chem. Soc. 2010, 132, 3264–3265. [Google Scholar] [CrossRef]
- Raghavan, A.; Charron, G.; Flexner, J.; Hang, H.C. Chemical probes for profiling fatty acid-associated proteins in living cells. Bioorg. Med. Chem. Lett. 2008, 18, 5982–5986. [Google Scholar] [CrossRef]
- Viertler, M.; Schittmayer, M.; Birner-Gruenberger, R. Activity based subcellular resolution imaging of lipases. Bioorg. Med. Chem. 2012, 20, 628–632. [Google Scholar] [CrossRef]
- Nasheri, N.; Joyce, M.; Rouleau, Y.; Yang, P.; Yao, S.; Tyrrell, D.L.; Pezacki, J.P. Modulation of fatty acid synthase enzyme activity and expression during hepatitis C virus replication. Chem. Biol. 2013, 20, 570–582. [Google Scholar] [CrossRef]
- Chang, J.W.; Cognetta, A.B.; Niphakis, M.J.; Cravatt, B.F. Proteome-wide reactivity profiling identifies diverse carbamate chemotypes tuned for serine hydrolase inhibition. ACS Chem. Biol. 2013, 8, 1590–1599. [Google Scholar] [CrossRef]
- Shannon, D.A.; Gu, C.; McLaughlin, C.J.; Kaiser, M.; van der Hoorn, R.A.; Weerapana, E. Sulfonyl fluoride analogues as activity-based probes for serine proteases. Chembiochem 2012, 13, 2327–2330. [Google Scholar] [CrossRef]
- Haedke, U.; Gotz, M.; Baer, P.; Verhelst, S.H. Alkyne derivatives of isocoumarins as clickable activity-based probes for serine proteases. Bioorg. Med. Chem. 2012, 20, 633–640. [Google Scholar] [CrossRef]
- Sabido, E.; Tarrago, T.; Giralt, E. Using peptidyl aldehydes in activity-based proteomics. Bioorg. Med. Chem. Lett. 2009, 19, 3752–3755. [Google Scholar] [CrossRef]
- Sabido, E.; Tarrago, T.; Niessen, S.; Cravatt, B.F.; Giralt, E. Activity-based probes for monitoring postproline protease activity. ChemBioChem 2009, 10, 2361–2366. [Google Scholar] [CrossRef]
- Wolf, E.V.; Zeissler, A.; Vosyka, O.; Zeiler, E.; Sieber, S.; Verhelst, S.H. A new class of rhomboid protease inhibitors discovered by activity-based fluorescence polarization. PLoS One 2013, 8, e72307. [Google Scholar]
- An, H.; Statsyuk, A.V. Development of activity-based probes for ubiquitin and ubiquitin-like protein signaling pathways. J. Am. Chem. Soc. 2013, 135, 16948–16962. [Google Scholar] [CrossRef]
- Ciechanover, A. Intracellular protein degradation: From a vague idea through the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Neurodegener. Dis. 2012, 10, 7–22. [Google Scholar] [CrossRef]
- Powers, J.C.; Asgian, J.L.; Ekici, O.D.; James, K.E. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem. Rev. 2002, 102, 4639–4750. [Google Scholar] [CrossRef]
- Greenbaum, D.; Medzihradszky, K.F.; Burlingame, A.; Bogyo, M. Epoxide electrophiles as activity-dependent cysteine protease profiling and discovery tools. Chem. Biol. 2000, 7, 569–581. [Google Scholar] [CrossRef]
- Kaschani, F.; Verhelst, S.H.; van Swieten, P.F.; Verdoes, M.; Wong, C.S.; Wang, Z.; Kaiser, M.; Overkleeft, H.S.; Bogyo, M.; van der Hoorn, R.A. Minitags for small molecules: Detecting targets of reactive small molecules in living plant tissues using “click chemistry”. Plant J. 2009, 57, 373–385. [Google Scholar] [CrossRef]
- Van der Linden, W.A.; Li, N.; Hoogendoorn, S.; Ruben, M.; Verdoes, M.; Guo, J.; Boons, G.J.; van der Marel, G.A.; Florea, B.I.; Overkleeft, H.S. Two-step bioorthogonal activity-based proteasome profiling using copper-free click reagents: A comparative study. Bioorg. Med. Chem. 2012, 20, 662–666. [Google Scholar] [CrossRef]
- Ovaa, H.; van Swieten, P.F.; Kessler, B.M.; Leeuwenburgh, M.A.; Fiebiger, E.; van den Nieuwendijk, A.M.; Galardy, P.J.; van der Marel, G.A.; Ploegh, H.L.; Overkleeft, H.S. Chemistry in living cells: Detection of active proteasomes by a two-step labeling strategy. Angew. Chem. Int. Ed. Engl. 2003, 42, 3626–3629. [Google Scholar] [CrossRef]
- Geurink, P.P.; van der Linden, W.A.; Mirabella, A.C.; Gallastegui, N.; de Bruin, G.; Blom, A.E.; Voges, M.J.; Mock, E.D.; Florea, B.I.; van der Marel, G.A.; et al. Incorporation of non-natural amino acids improves cell permeability and potency of specific inhibitors of proteasome trypsin-like sites. J. Med. Chem. 2013, 56, 1262–1275. [Google Scholar] [CrossRef]
- Bicker, K.L.; Thompson, P.R. The protein arginine deiminases: Structure, function, inhibition, and disease. Biopolymers 2013, 99, 155–163. [Google Scholar] [CrossRef]
- Luo, Y.; Knuckley, B.; Bhatia, M.; Pellechia, P.J.; Thompson, P.R. Activity-based protein profiling reagents for protein arginine deiminase 4 (PAD4): Synthesis and in vitro evaluation of a fluorescently labeled probe. J. Am. Chem. Soc. 2006, 128, 14468–14469. [Google Scholar] [CrossRef]
- Slack, J.L.; Causey, C.P.; Luo, Y.; Thompson, P.R. Development and use of clickable activity based protein profiling agents for protein arginine deiminase 4. ACS Chem. Biol. 2011, 6, 466–476. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, S.; Fast, W. A click chemistry mediated in vivo activity probe for dimethylarginine dimethylaminohydrolase. J. Am. Chem. Soc. 2009, 131, 15096–15097. [Google Scholar] [CrossRef]
- Denisov, I.G.; Makris, T.M.; Sligar, S.G.; Schlichting, I. Structure and chemistry of cytochrome P450. Chem. Rev. 2005, 105, 2253–2277. [Google Scholar] [CrossRef]
- Wright, A.T.; Song, J.D.; Cravatt, B.F. A suite of activity-based probes for human cytochrome P450 enzymes. J. Am. Chem. Soc. 2009, 131, 10692–10700. [Google Scholar] [CrossRef]
- Severi, E.; Hood, D.W.; Thomas, G.H. Sialic acid utilization by bacterial pathogens. Microbiology 2007, 153, 2817–2822. [Google Scholar] [CrossRef]
- Tsai, C.S.; Yen, H.Y.; Lin, M.I.; Tsai, T.I.; Wang, S.Y.; Huang, W.I.; Hsu, T.L.; Cheng, Y.S.; Fang, J.M.; Wong, C.H. Cell-permeable probe for identification and imaging of sialidases. Proc. Natl. Acad. Sci. USA 2013, 110, 2466–2471. [Google Scholar] [CrossRef]
- Overall, C.M.; Lopez-Otin, C. Strategies for MMP inhibition in cancer: Innovations for the post-trial era. Nat. Rev. Cancer 2002, 2, 657–672. [Google Scholar] [CrossRef]
- Sieber, S.A.; Niessen, S.; Hoover, H.S.; Cravatt, B.F. Proteomic profiling of metalloprotease activities with cocktails of active-site probes. Nat. Chem. Biol. 2006, 2, 274–281. [Google Scholar]
- You, A.; Tong, J.K.; Grozinger, C.M.; Schreiber, S.L. CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc. Natl. Acad. Sci. USA 2001, 98, 1454–1458. [Google Scholar] [CrossRef]
- Salisbury, C.M.; Cravatt, B.F. Optimization of activity-based probes for proteomic profiling of histone deacetylase complexes. J. Am. Chem. Soc. 2008, 130, 2184–2194. [Google Scholar] [CrossRef]
- Salisbury, C.M.; Cravatt, B.F. Activity-based probes for proteomic profiling of histone deacetylase complexes. Proc. Natl. Acad. Sci. USA 2007, 104, 1171–1176. [Google Scholar] [CrossRef]
- Kalesh, K.A.; Sim, D.S.; Wang, J.; Liu, K.; Lin, Q.; Yao, S.Q. Small molecule probes that target Abl kinase. Chem. Commun. 2010, 46, 1118–1120. [Google Scholar]
- Tantama, M.; Lin, W.C.; Licht, S. An activity-based protein profiling probe for the nicotinic acetylcholine receptor. J. Am. Chem. Soc. 2008, 130, 15766–15767. [Google Scholar] [CrossRef]
- De Strooper, B.; Vassar, R.; Golde, T. The secretases: Enzymes with therapeutic potential in Alzheimer disease. Nat. Rev. Neurol. 2010, 6, 99–107. [Google Scholar] [CrossRef]
- Zettl, H.; Weggen, S.; Schneider, P.; Schneider, G. Exploring the chemical space of gamma-secretase modulators. Trends Pharmacol. Sci. 2010, 31, 402–410. [Google Scholar] [CrossRef]
- Pozdnyakov, N.; Murrey, H.E.; Crump, C.J.; Pettersson, M.; Ballard, T.E.; Am Ende, C.W.; Ahn, K.; Li, Y.M.; Bales, K.R.; Johnson, D.S. gamma-Secretase modulator (GSM) photoaffinity probes reveal distinct allosteric binding sites on presenilin. J. Biol. Chem. 2013, 288, 9710–9720. [Google Scholar] [CrossRef]
- Crump, C.J.; Johnson, D.S.; Li, Y.M. Development and mechanism of gamma-secretase modulators for Alzheimer’s disease. Biochemistry 2013, 52, 3197–3216. [Google Scholar] [CrossRef]
- Crump, C.J.; am Ende, C.W.; Ballard, T.E.; Pozdnyakov, N.; Pettersson, M.; Chau, D.M.; Bales, K.R.; Li, Y.M.; Johnson, D.S. Development of clickable active site-directed photoaffinity probes for gamma-secretase. Bioorg. Med. Chem. Lett. 2012, 22, 2997–3000. [Google Scholar] [CrossRef]
- Cravatt, B.F.; Simon, G.M.; Yates, J.R., 3rd. The biological impact of mass-spectrometry-based proteomics. Nature 2007, 450, 991–1000. [Google Scholar] [CrossRef]
- Weerapana, E.; Speers, A.E.; Cravatt, B.F. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)—A general method for mapping sites of probe modification in proteomes. Nat. Protoc. 2007, 2, 1414–1425. [Google Scholar] [CrossRef]
- Speers, A.E.; Cravatt, B.F. A tandem orthogonal proteolysis strategy for high-content chemical proteomics. J. Am. Chem. Soc. 2005, 127, 10018–10019. [Google Scholar] [CrossRef]
- Weerapana, E.; Wang, C.; Simon, G.M.; Richter, F.; Khare, S.; Dillon, M.B.; Bachovchin, D.A.; Mowen, K.; Baker, D.; Cravatt, B.F. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 2010, 468, 790–795. [Google Scholar] [CrossRef]
- Qian, Y.; Martell, J.; Pace, N.J.; Ballard, T.E.; Johnson, D.S.; Weerapana, E. An isotopically tagged azobenzene-based cleavable linker for quantitative proteomics. ChemBioChem 2013, 14, 1410–1414. [Google Scholar] [CrossRef]
- Weerapana, E.; Simon, G.M.; Cravatt, B.F. Disparate proteome reactivity profiles of carbon electrophiles. Nat. Chem. Biol. 2008, 4, 405–407. [Google Scholar] [CrossRef]
- Wang, C.; Weerapana, E.; Blewett, M.M.; Cravatt, B.F. A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat. Methods 2014, 11, 79–85. [Google Scholar]
- Pace, N.J.; Weerapana, E. A competitive chemical-proteomic platform to identify zinc-binding cysteines. ACS Chem. Biol. 2013. [Google Scholar] [CrossRef]
- Yang, J.; Chaurand, P.; Norris, J.L.; Porter, N.A.; Caprioli, R.M. Activity-based probes linked with laser-cleavable mass tags for signal amplification in imaging mass spectrometry: Analysis of serine hydrolase enzymes in mammalian tissue. Anal. Chem. 2012, 84, 3689–3695. [Google Scholar] [CrossRef]
- Yan, X.W.; Luo, Y.C.; Zhang, Z.B.; Li, Z.X.; Luo, Q.; Yang, L.M.; Zhang, B.; Chen, H.F.; Bai, P.M.; Wang, Q.Q. Europium-labeled activity-based probe through click chemistry: Absolute serine protease quantification using 153Eu isotope dilution ICP/MS. Angew. Chem. Int. Ed. 2012, 51, 3358–3363. [Google Scholar] [CrossRef]
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Martell, J.; Weerapana, E. Applications of Copper-Catalyzed Click Chemistry in Activity-Based Protein Profiling. Molecules 2014, 19, 1378-1393. https://doi.org/10.3390/molecules19021378
Martell J, Weerapana E. Applications of Copper-Catalyzed Click Chemistry in Activity-Based Protein Profiling. Molecules. 2014; 19(2):1378-1393. https://doi.org/10.3390/molecules19021378
Chicago/Turabian StyleMartell, Julianne, and Eranthie Weerapana. 2014. "Applications of Copper-Catalyzed Click Chemistry in Activity-Based Protein Profiling" Molecules 19, no. 2: 1378-1393. https://doi.org/10.3390/molecules19021378
APA StyleMartell, J., & Weerapana, E. (2014). Applications of Copper-Catalyzed Click Chemistry in Activity-Based Protein Profiling. Molecules, 19(2), 1378-1393. https://doi.org/10.3390/molecules19021378



