Biochemical Characteristics of Three Laccase Isoforms from the Basidiomycete Pleurotus nebrodensis
Abstract
:1. Introduction
2. Results
2.1. Purification of Laccases

| Chromatographic Method | Chromatographic Fraction by Steps | Yield (mg) | Total Activity (U) a | Specific Activity (U/mg) b | Recovery of Activity (%) | Purification Fold c |
|---|---|---|---|---|---|---|
| None | Crude extract | 23040 | 2853 | 0.12 | 100 | 1 |
| DEAE-cellulose anion exchange | D2 (Lac 1 and Lac2) | 3200 | 1323 | 0.41 | 46.38 | 3.3 |
| D3 (Lac3) | 175 | 689 | 3.94 | 24.14 | 32 | |
| CM-cellulose cation exchange | D2C1 (Lac1) | 62 | 33 | 0.53 | 1.16 | 4.2 |
| D2C2 (Lac2) | 32 | 297 | 9.31 | 10.42 | 75 | |
| D3C (Lac3) | 29 | 64 | 2.24 | 2.24 | 18 | |
| Q-Sepharose anion exchange | D2C1Q (Lac1) | 31 | 18.6 | 0.59 | 0.65 | 4.8 |
| D2C2Q (Lac2) | 4.2 | 49.8 | 11.85 | 1.74 | 96 | |
| D3CQ (Lac3) | 9.6 | 27 | 2.81 | 0.95 | 23 | |
| Superdex 75 size exclusion | D2C1QS (Lac1) | 1.58 | 10.8 | 6.86 | 0.38 | 55 |
| D2C2QS (Lac2) | 1.13 | 25.6 | 22.76 | 0.90 | 183 | |
| D3CQS (Lac3) | 0.13 | 22.4 | 171 | 0.79 | 1379 |

2.2. Effects of pH and Temperature on Laccase Activity and Stability
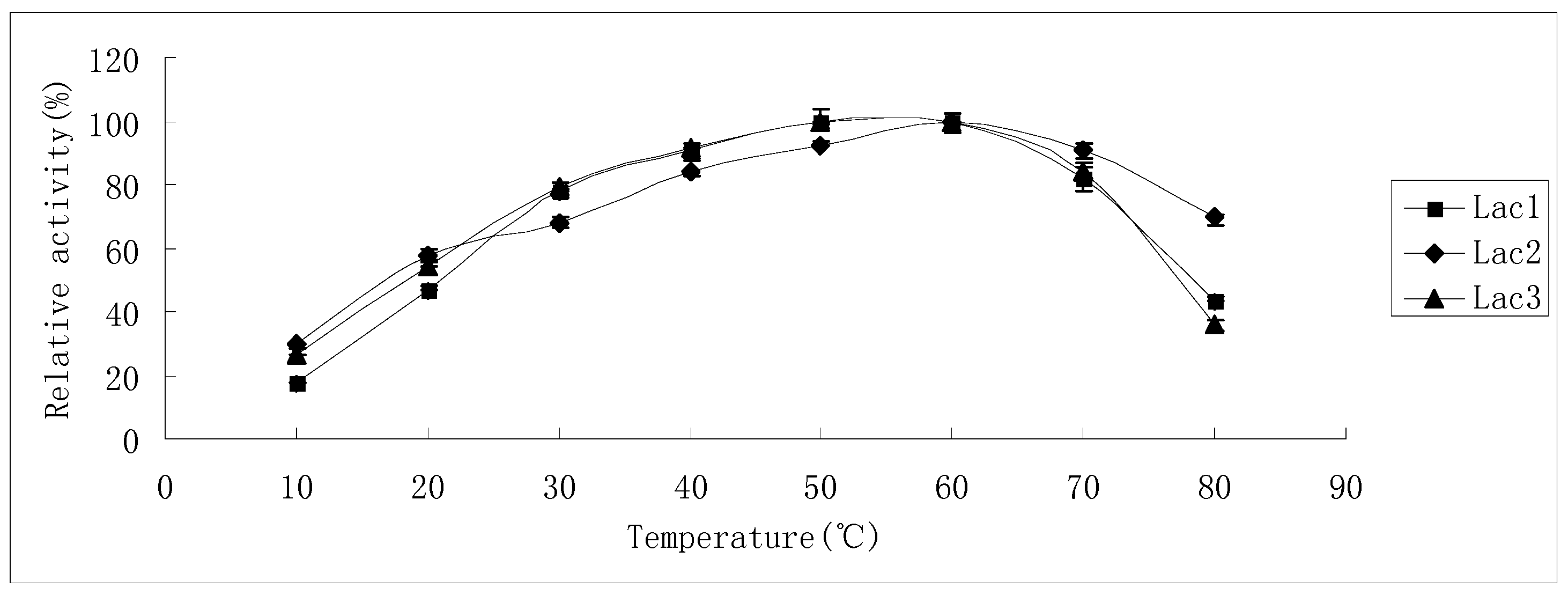




2.3. Substrate Specificity of Isolated Laccases
| Substrate | Wavelength (nm) | Relative Laccase Activity (%) | ||
|---|---|---|---|---|
| Lac1 | Lac2 | Lac3 | ||
| ABTS | 420 | 100 ± 1.0 | 100 ± 0.0 | 100 ± 1.5 |
| Guaiacol | 460 | 16 ± 0.08 | 24 ± 0.08 | 100 ± 0.04 |
| Dimethylphthalate | 470 | 22.83 ± 2.17 | 100 ± 3.0 | 92.39 ± 3.26 |
| Syringaldazine | 525 | 100 ± 4.23 | 89.44 ± 4.93 | 84.51 ± 1.41 |
| l-DOPA | 470 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Ferulic acid | 318 | 72.73 ± 3.64 | 3.64 ± 0.0 | 100 ± 1.82 |
| Caffeic acid | 318 | 34.55 ± 1.82 | 12.5 ± 0.0 | 100 ± 1.82 |
| Vanillic acid | 261 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| p-Coumaric acid | 318 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Hydroquinone | 289 | 25 ± 1.56 | 6.25 ± 0.0 | 100 ± 3.125 |
| Catechol | 276 | 100 ± 0.14 | 92.86 ± 0.56 | 42.86 ± 0.28 |
| p-Phenylenediamine | 250 | 18.2 ± 0.0 | 54.55 ± 0.27 | 100 ± 0.09 |
2.4. Kinetic Constants for Laccases
| Laccase Isoforms | Km (mM) | Kcat (min−1) | Kcat/Km (min−1·mM−1) |
|---|---|---|---|
| Lac1 | 0.16 | 0.958 | 5.99 |
| Lac2 | 0.55 | 1.65 | 3.00 |
| Lac3 | 0.104 | 0.264 | 2.54 |
2.5. Peptides Identified by Gel-Fractionation LC-LTQ-Orbitrap-MS
| Laccaes Isoforms | |||
|---|---|---|---|
| Lac1 | Lac2 | Lac3 | |
| Peptides | RNDVVSPDGFERR | KVIQPDGFSRS | KGDNFQLNVVNQLSDTTMLKD |
| RRAITVNGIFPGTPVILQKN | SAVLAGGSYPGPLIKG | RYAGGPTSPLAIINVESNKRY | |
| KNDKVQINTINELTDPGMRR | RLYDVDDESTVLTVGDWYHAPSLSLTGVPHPDSTLFNGLGRS | RANPNLGSTGFDGGINSAILRY | |
| RSTSIHWHGLFQHKT | SLNGPASPLYVMNVVK | ||
| RYKGAPAVEPTTVATTGGHKL | RYSLVLNANQAVGNYWIRA | ||
| RKPQDFLPSEQVIILPANKL | ANPNSGDPGFANQMNSAILRY | ||
| RTSNSDVVNLVNPPRR | REYNLRPLIKK | ||
| RDVLPINGGNTTFRF | RDAHDLAPAGSIYDIKL | ||
| RTLCPAYDGLAPEFQ | LGDVVEITMPALVFAGPHPLHLQWHTFAVVRS | ||
| SAGSSTYNYENPVRRD | |||
| DVVSIGDDPTDNVTIRF | |||
3. Discussion
4. Experimental Section
4.1. Chemical Reagents and Microorganism
4.2. Enzyme Activity Assay
4.3. Purification of Laccase isoforms
4.4. Biochemical Characterization of the Purified Laccase Isoforms
4.4.1. Effects of Temperature and pH on Laccase Activity and Stability
4.4.2. Assay for Substrate Specificity
4.4.3. Kinetic Constants of Laccase Isoforms
4.5. Molecular Weight Determination and Peptide Identification with Gel-Fractionation LC-LTQ-Orbitrap-MS
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jiang, M.; Ten, Z.; Ding, S. Decolorization of synthetic dyes by crude and purified laccases from Coprinus comatus grown under different cultures: The role of major isoenzyme in dyes decolorization. Appl. Biochem. Biotechnol. 2013, 169, 660–672. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.M.; Staples, R.C. Laccase: New functions for an old enzyme. Phytochemistry 2002, 60, 551–565. [Google Scholar] [CrossRef]
- Ranocha, P.; McDougall, G.; Hawkins, S.; Sterjiades, R.; Borderies, G.; Stewart, D.; Cabanes-Macheteau, M.; Boudet, A.M.; Goffner, D. Biochemical characterization, molecular cloning and expression of laccases—A divergent gene family—In poplar. Eur. J. Biochem. 1999, 259, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Viswanath, B.; Rajesh, B.; Janardhan, A.; Kumar, A.P.; Narasimha, G. Fungal laccases and their applications in bioremediation. Enzym. Res. 2014, 2014, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Pant, D.; Adholeya, A. Concentration of fungal ligninolytic enzymes by ultrafiltration and their use in distillery effluent decolorization. World J. Microbiol. Biotechnol. 2009, 25, 1793–1800. [Google Scholar] [CrossRef]
- Baldrian, P. Fungal laccases—Occurrence and properties. FEMS Microbiol. Rev. 2006, 30, 215–242. [Google Scholar] [CrossRef] [PubMed]
- Collins, P.J.; Dobson, A. Regulation of laccase gene transcription in Trametes versicolor. Appl. Environ. Microbiol. 1997, 63, 3444–3450. [Google Scholar] [PubMed]
- Giardina, P.; Palmieri, G.; Scaloni, A.; Fontanella, B.; Faraco, V.; Cennamo, G.; Sannia, G. Protein and gene structure of a blue laccase from Pleurotus ostreatus1. Biochem. J. 1999, 341, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, G.; Cennamo, G.; Faraco, V.; Amoresano, A.; Sannia, G.; Giardina, P. Atypical laccase isoenzymes from copper supplemented Pleurotus ostreatus cultures. Enzym. Microb. Technol. 2003, 33, 220–230. [Google Scholar] [CrossRef]
- Palmieri, G.; Giardina, P.; Bianco, C.; Scaloni, A.; Capasso, A.; Sannia, G. A novel white laccase from Pleurotus ostreatus. J. Biol. Chem. 1997, 272, 31301–31307. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, G.; Giardina, P.; Bianco, C.; Fontanella, B.; Sannia, G. Copper induction of laccase isoenzymes in the ligninolytic fungus Pleurotus ostreatus. Appl. Environ. Microbiol. 2000, 66, 920–924. [Google Scholar] [CrossRef] [PubMed]
- Couto, S.R.; Herrera, J.L.T. Industrial and biotechnological applications of laccases: A review. Biotechnol. Adv. 2006, 24, 500–513. [Google Scholar] [CrossRef] [PubMed]
- Torres-Duarte, C.; Roman, R.; Tinoco, R.; Vazquez-Duhalt, R. Halogenated pesticide transformation by a laccase–mediator system. Chemosphere 2009, 77, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Pickard, M.A.; Roman, R.; Tinoco, R.; Vazquez-Duhalt, R. Polycyclic aromatic hydrocarbon metabolism by white rot fungi and oxidation by Coriolopsis gallica UAMH 8260 laccase. Appl. Environ. Microbiol. 1999, 65, 3805–3809. [Google Scholar] [PubMed]
- Jia, J.; Zhang, S.; Wang, P.; Wang, H. Degradation of high concentration 2,4-dichlorophenol by simultaneous photocatalytic–enzymatic process using TiO2/UV and laccase. J. Hazard. Mater. 2012, 205, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Tian, G.-T.; Zhang, G.-Q.; Wang, H.-X.; Ng, T.B. Purification and characterization of a novel laccase from the mushroom Pleurotus nebrodensis. Acta Biochim. Pol. 2012, 59, 407–412. [Google Scholar] [PubMed]
- Das, N.; Chakraborty, T.K.; Mukherjee, M. Purification and characterization of laccase-1 from Pleurotus florida. Folia Microbiol. 2000, 45, 447–451. [Google Scholar] [CrossRef]
- Marques de Souza, C.G.; Peralta, R.M. Purification and characterization of the main laccase produced by the white-rot fungus Pleurotus pulmonarius on wheat bran solid state medium. J. Basic Microbiol. 2003, 43, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Sondhi, S.; Sharma, P.; Saini, S.; Puri, N.; Gupta, N. Purification and characterization of an extracellular, thermo-alkali-stable, metal tolerant laccase from Bacillus tequilensis SN4. PLoS ONE 2014, 9, e96951. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, S.; Diao, H.; Zhao, H.; Zhu, X.; Lu, F.; Lu, Z. Purification and characterization of a temperature-and pH-stable laccase from the spores of Bacillus vallismortis FMB-103 and its application in the degradation of malachite green. J. Agric. Food Chem. 2013, 61, 5468–5473. [Google Scholar] [CrossRef] [PubMed]
- Leonowicz, A.; Trojanowski, J.; Orlicz, B. Induction of laccase in Basidiomycetes: Apparent activity of the inducible and constitutive forms of the enzyme with phenolic substrates. Acta Biochim. Pol. 1978, 25, 369–378. [Google Scholar] [PubMed]
- Murugesan, K.; Arulmani, M.; Nam, I.-H.; Kim, Y.-M.; Chang, Y.-S.; Kalaichelvan, P.T. Purification and characterization of laccase produced by a white rot fungus Pleurotus sajor-caju under submerged culture condition and its potential in decolorization of azo dyes. Appl. Microbiol. Biotechnol. 2006, 72, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Karp, S.G.; Faraco, V.; Amore, A.; Birolo, L.; Giangrande, C.; Soccol, V.T.; Pandey, A.; Soccol, C.R. Characterization of laccase isoforms produced by Pleurotus ostreatus in solid state fermentation of sugarcane bagasse. Bioresour. Technol. 2012, 114, 735–739. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, P.; Hari, R.; Chakraborty, B.; Mandal, B.; Naskar, S.; Das, N. Isolation, culture optimization and physico-chemical characterization of laccase enzyme from Pleurotus fossulatus. Pak. J. Biol. Sci. 2014, 17, 173–181. [Google Scholar] [PubMed]
- Eichlerová, I.; Homolka, L.; Nerud, F. Decolorization of orange G by Pleurotus ostreatus monokaryotic isolates with different laccase activity. Folia Microbiol. 2003, 48, 775–779. [Google Scholar] [CrossRef]
- Reiss, R.; Ihssen, J.; Thöny-Meyer, L. Bacillus pumilus laccase: A heat stable enzyme with a wide substrate spectrum. BMC Biotechnol. 2011, 11. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, E.V.; Pegasova, T.V.; Gavrilova, V.P.; Landesman, E.O.; Koroleva, O.V. Extracellular laccases from Cerrena unicolor 059, Cerrena unicolor 0784, and Pleurotus oastreatus 0432: A comparative assay. Appl. Biochem. Microbiol. 2003, 39, 375–381. [Google Scholar] [CrossRef]
- Solomon, E.I.; Sundaram, U.M.; Machonkin, T.E. Multicopper oxidases and oxygenases. Chem. Rev. 1996, 96, 2563–2606. [Google Scholar] [CrossRef] [PubMed]
- Almansa, E.; Kandelbauer, A.; Pereira, L.; Cavaco-Paulo, A.; Guebitz, G.M. Influence of structure on dye degradation with laccase mediator systems. Biocatal. Biotransformation 2004, 22, 315–324. [Google Scholar] [CrossRef]
- Kandelbauer, A.; Erlacher, A.; Cavaco-Paulo, A.; Guebitz, G.M. Laccase-catalyzed decolorization of the synthetic azo-dye diamond black PV 200 and of some structurally related derivatives. Biocatal. Biotransform. 2004, 22, 331–339. [Google Scholar] [CrossRef] [Green Version]
- Tadesse, M.A.; D'Annibale, A.; Galli, C.; Gentili, P.; Sergi, F. An assessment of the relative contributions of redox and steric issues to laccase specificity towards putative substrates. Org. Biomol. Chem. 2008, 6, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Kulys, J.J.; Duke, K.; Li, K.; Krikstopaitis, K.; Deussen, H.-J.W.; Abbate, E.; Galinyte, V.; Schneider, P. Redox chemistry in laccase-catalyzed oxidation of N-hydroxy compounds. Appl. Environ. Microbiol. 2000, 66, 2052–2056. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Xu, F.; Eriksson, K.-E.L. Comparison of fungal laccases and redox mediators in oxidation of a nonphenolic lignin model compound. Appl. Environ. Microbiol. 1999, 65, 2654–2660. [Google Scholar] [PubMed]
- Lisova, Z.A.; Lisov, A.V.; Leontievsky, A.A. Two laccase isoforms of the basidiomycete Cerrena unicolor VKMF-3196. Induction, isolation and properties. J. Basic Microbiol. 2010, 50, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Zheng, F.; Long, L.; Wang, J.; Ding, S. Engineering the expression and characterization of two novel laccase isoenzymes from Coprinus comatus in Pichia pastoris by fusing an additional ten amino acids tag at N-terminus. PLoS ONE 2014, 9, e93912. [Google Scholar] [CrossRef] [PubMed]
- Sherif, M.; Waung, D.; Korbeci, B.; Mavisakalyan, V.; Flick, R.; Brown, G.; Abou-Zaid, M.; Yakunin, A.F.; Master, E.R. Biochemical studies of the multicopper oxidase (small laccase) from Streptomyces coelicolor using bioactive phytochemicals and site-directed mutagenesis. Microb. Biotechnol. 2013, 6, 588–597. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhu, Y.; Wang, P.; Zhu, L.; Zheng, J.; Li, R.; Ruan, L.; Peng, D.; Sun, M. Complete genome sequence of Bacillus subtilis BSn5, an endophytic bacterium of Amorphophallus konjac with antimicrobial activity for the plant pathogen Erwinia carotovora subsp. carotovora. J. Bacteriol. 2011, 193, 2070–2071. [Google Scholar] [CrossRef] [PubMed]
- Sitarz, A.K.; Mikkelsen, J.D.; Højrup, P.; Meyer, A.S. Identification of a laccase from Ganoderma lucidum CBS 229.93 having potential for enhancing cellulase catalyzed lignocellulose degradation. Enzym. Microb. Technol. 2013, 53, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, E.; Ruiz-Duenas, F.; Kooistra, R.; Ram, A.; Martinez, A. Isolation of two laccase genes from the white-rot fungus Pleurotus eryngii and heterologous expression of the pel3 encoded protein. J. Biotechnol. 2008, 134, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K.; Favre, M. Maturation of the head of bacteriophage T4: I. DNA packaging events. J. Mol. Biol. 1973, 80, 575–599. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds are not available from the authors.
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, X.; Tian, G.; Zhao, Y.; Zhao, L.; Wang, H.; Ng, T.B. Biochemical Characteristics of Three Laccase Isoforms from the Basidiomycete Pleurotus nebrodensis. Molecules 2016, 21, 203. https://doi.org/10.3390/molecules21020203
Yuan X, Tian G, Zhao Y, Zhao L, Wang H, Ng TB. Biochemical Characteristics of Three Laccase Isoforms from the Basidiomycete Pleurotus nebrodensis. Molecules. 2016; 21(2):203. https://doi.org/10.3390/molecules21020203
Chicago/Turabian StyleYuan, Xianghe, Guoting Tian, Yongchang Zhao, Liyan Zhao, Hexiang Wang, and Tzi Bun Ng. 2016. "Biochemical Characteristics of Three Laccase Isoforms from the Basidiomycete Pleurotus nebrodensis" Molecules 21, no. 2: 203. https://doi.org/10.3390/molecules21020203




