Effect of Phyllanthus amarus Extract on 5-Fluorouracil-Induced Perturbations in Ribonucleotide and Deoxyribonucleotide Pools in HepG2 Cell Line
Abstract
:1. Introduction
2. Results and Discussion
2.1. Standarization of PHA Extract
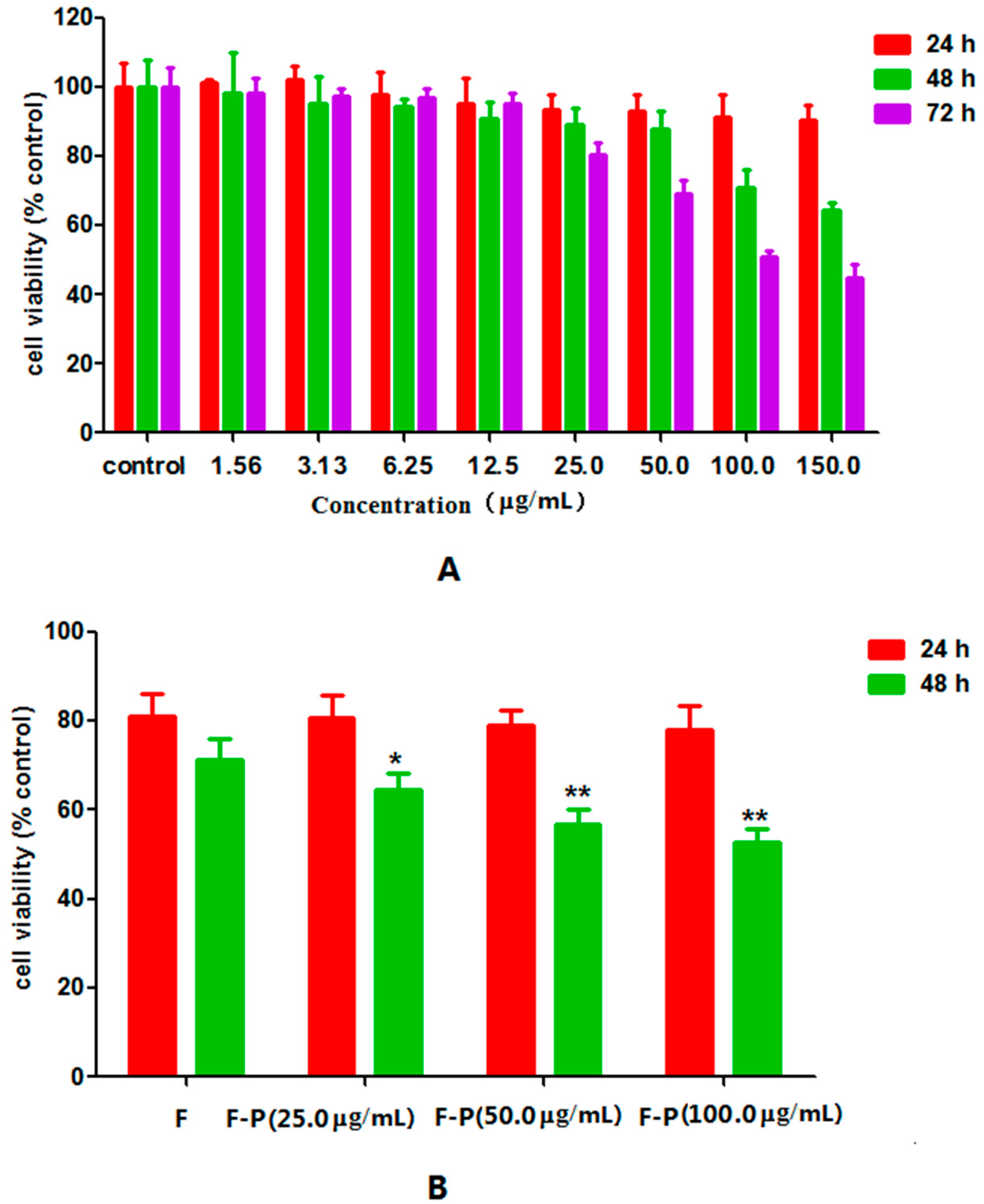
2.2. Effect of PHA Extract on Growth of HepG2 Cells
2.3. Multivariate Statistical Analysis
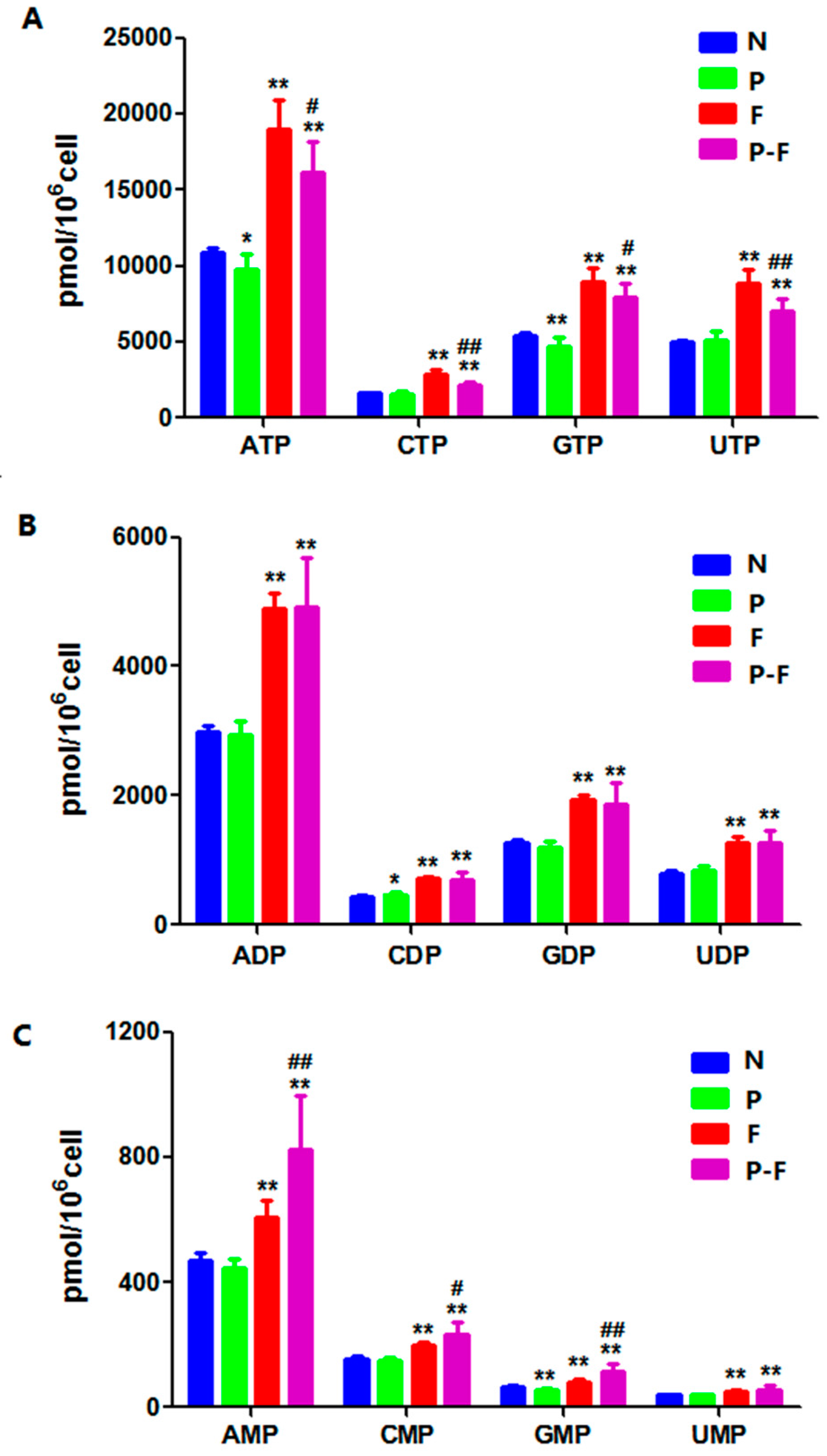
2.4. Effect of PHA Extract Incubation on RNs and dRNs Pool Sizes
2.5. PHA extract Induced Cell Cycle Arrest in HepG2 Cells
3. Materials and Methods
3.1. Materials
3.2. Preparation of PHA Extract
3.3. LC/MS/MS Assay
3.4. Cell Culture
3.5. Preparation of Cell Pellets
3.6. MTT Assay
3.7. Cell Cycle Analysis
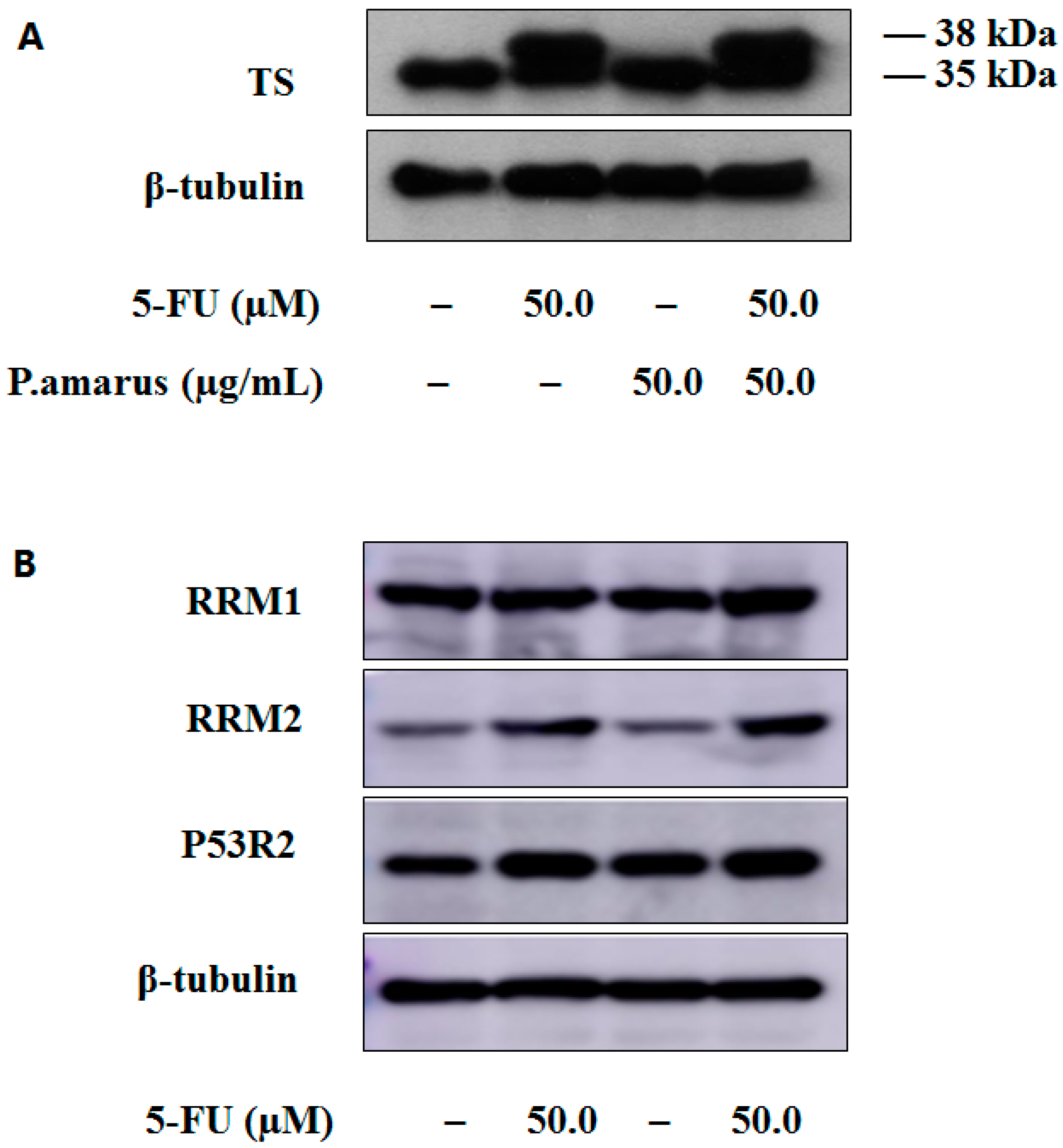
3.8. Western Blot Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jordheim, L.P.; Durantel, D.; Zoulim, F.; Dumontet, C. Advances in the development of nucleoside and nucleotide analogs for cancer and viral diseases. Nat. Rev. Drug Discov. 2013, 12, 447–464. [Google Scholar] [CrossRef] [PubMed]
- Krupp, G. Enzymatic RNA synthesis and RNase P. Evolutionary aspects. Mol. Biol. Rep. 1995, 22, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Van Rompay, A.R.; Johansson, M.; Karlsson, A. Phosphorylation of nucleosides and nucleoside analogs by mammalian nucleoside monophosphate kinases. Pharmacol. Ther. 2000, 87, 189–198. [Google Scholar] [CrossRef]
- Houghton, J.A.; Tillman, D.M.; Harwood, F.G. Ratio of 2′-deoxyadenosine-5′-triphosphate/thymidine-5′-triphosphate influences the commitment of human colon carcinoma cells to thymineless death. Clin. Cancer Res. 1995, 1, 723–730. [Google Scholar] [PubMed]
- Mitrovski, B.; Pressacco, J.; Mandelbaum, S.; Erlichman, C. Biochemical effects of folate-based inhibitors of thymidylate synthase in MGH-U1 cells. Cancer Chemother. Pharmacol. 1994, 35, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Marsoni, S. Efficacy of adjuvant fluorouracil and folinic acid in colon cancer. International Multicentre Pooled Analysis of Colon Cancer Trials (IMPACT) investigators. Lancet 1995, 345, 939–944. [Google Scholar]
- Douillard, J.Y.; Cunningham, D.; Roth, A.D.; Navarro, M.; James, R.D.; Karasek, P.; Jandik, P.; Iveson, T.; Carmichael, J.; Alakl, M.; et al. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: A multicentre randomised trial. Lancet 2000, 355, 1041–1047. [Google Scholar] [CrossRef]
- Johnston, P.G.; Kaye, S. Capecitabine: A novel agent for the treatment of solid tumors. Anticancer Drugs 2001, 12, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-Fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Kinouchi, M.; Ishida, K.; Fujibuchi, W.; Naitoh, T.; Ogawa, H.; Ando, T.; Yazaki, N.; Watanabe, K.; Haneda, S.; et al. 5-Fu metabolism in cancer and orally-administrable 5-fu drugs. Cancers (Basel) 2010, 2, 1717–1730. [Google Scholar] [CrossRef] [PubMed]
- Qi, F.H.; Li, A.Y.; Inagaki, Y.; Gao, J.J.; Li, J.J.; Kokudo, N.; Li, X.K.; Tang, W. Chinese herbal medicines as adjuvant treatment during chemo- or radio-therapy for cancer. Biosci. Trends 2010, 4, 297–307. [Google Scholar] [PubMed]
- Konkimalla, V.B.; Efferth, T. Evidence-based Chinese medicine for cancer therapy. J. Ethnopharmacol. 2008, 116, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Roengrit, T.; Wannanon, P.; Prasertsri, P.; Kanpetta, Y.; Sripanidkulchai, B.O.; Wattanathorn, J.; Leelayuwat, N. Antioxidant effect of Phyllanthus amarus after moderate-intensity exercise in sedentary males: A randomized crossover (double-blind) study. J. Phys. Ther. Sci. 2015, 27, 1181–1186. [Google Scholar] [CrossRef] [PubMed]
- Calixto, J.B.; Santos, A.R.; Cechinel Filho, V.; Yunes, R.A. A review of the plants of the genus Phyllanthus: Their chemistry, pharmacology, and therapeutic potential. Med. Res. Rev. 1998, 18, 225–258. [Google Scholar] [CrossRef]
- Patel, J.R.; Tripathi, P.; Sharma, V.; Chauhan, N.S.; Dixit, V.K. Phyllanthus amarus: Ethnomedicinal uses, phytochemistry and pharmacology: A review. J. Ethnopharmacol. 2011, 138, 286–313. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.Q.; Jaganath, I.B.; Sekaran, S.D. Phyllanthus spp. induces selective growth inhibition of PC-3 and MeWo human cancer cells through modulation of cell cycle and induction of apoptosis. PLoS ONE 2010, 5, e12644. [Google Scholar] [CrossRef] [PubMed]
- Islam, A.; Selvan, T.; Mazumder, U.; Gupta, M.; Ghosal, S. Antitumour effect of phyllanthin and hypophyllanthin from Phyllanthus amarus against Ehrlich ascites carcinoma in mice. Pharmacologyonline 2008, 2, 796–807. [Google Scholar]
- Blumberg, B.S.; Millman, I.; Venkateswaran, P.S.; Thyagarajan, S.P. Hepatitis B virus and primary hepatocellular carcinoma: Treatment of HBV carriers with Phyllanthus amarus. Vaccine 1990, 8, S86–S92. [Google Scholar] [CrossRef]
- Ott, M.; Thyagarajan, S.P.; Gupta, S. Phyllanthus amarus suppresses hepatitis B virus by interrupting interactions between HBV enhancer I and cellular transcription factors. Eur. J. Clin. Investig. 1997, 27, 908–915. [Google Scholar] [CrossRef]
- Rajeshkumar, N.V.; Joy, K.L.; Kuttan, G.; Ramsewak, R.S.; Nair, M.G.; Kuttan, R. Antitumour and anticarcinogenic activity of Phyllanthus amarus extract. J. Ethnopharmacol. 2002, 81, 17–22. [Google Scholar] [CrossRef]
- Taesotikul, T.; Dumrongsakulchai, W.; Wattanachai, N.; Navinpipat, V.; Somanabandhu, A.; Tassaneeyakul, W. Inhibitory effects of Phyllanthus amarus and its major lignans on human microsomal cytochrome P450 activities: Evidence for CYP3A4 mechanism-based inhibition. Drug Metab. Pharmacokinet. 2011, 26, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Colombo, R.; De L, B.; Andrea, N.; Teles, H.L.; Silva, G.H.; Bomfim, G.C.; Burgos, R.C.; Cavalheiro, A.J.; Da Silva Bolzani, V.; Silva, D.H.S. Validated HPLC method for the standardization of Phyllanthus niruri (herb and commercial extracts) using corilagin as a phytochemical marker. Biomedical. Chromatogr. 2009, 23, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Chen, Q.; Wang, C.; Qiu, H.; Liu, B.; Jiang, Z.H.; Zhang, W. Comparison of two exploratory data analysis methods for classification of Phyllanthus chemical fingerprint: Unsupervised vs. supervised pattern recognition technologies. Anal. Bioanal. Chem. 2015, 407, 1389–1401. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.R.; Chen, Q.Q.; Lam, C.W.K.; Wang, C.Y.; Wong, V.K.W.; Xu, F.G.; Jiang, Z.H.; Zhang, W. Application of artificial neural network to investigate the effects of 5-fluorouracil on ribonucleotides and deoxyribonucleotides in HepG2 cells. Sci. Rep. 2015, 5, 16861. [Google Scholar] [CrossRef] [PubMed]
- Kakito, H.; Ohkubo, T.; Kagawa, Y.; Inagaki, S.; Sumida, K.; Ooi, K.; Higashigawa, M.; Sakurai, M. Effects of 5-fluorouracil and a combination of tegafur and uracil (UFT) on nucleotide metabolism in L1210 ascites tumor. Cancer Investig. 1993, 11, 530–533. [Google Scholar] [CrossRef] [PubMed]
- Chu, E.; Koeller, D.M.; Johnston, P.G.; Zinn, S.; Allegra, C.J. Regulation of thymidylate synthase in human colon cancer cells treated with 5-fluorouracil and interferon-gamma. Mol. Pharmacol. 1993, 43, 527–533. [Google Scholar] [PubMed]
- Maley, G.F.; Maley, F. The Purification and Properties of Deoxycytidylate Deaminase from Chick Embryo Extracts. J. Biol. Chem. 1964, 239, 168–176. [Google Scholar] [PubMed]
- Mah, V.; Alavi, M.; Marquez-Garban, D.C.; Maresh, E.L.; Kim, S.R.; Horvath, S.; Bagryanova, L.; Huerta-Yepez, S.; Chia, D.; Pietras, R.; et al. Ribonucleotide reductase subunit M2 predicts survival in subgroups of patients with non-small cell lung carcinoma: Effects of gender and smoking status. PLoS ONE 2015, 10, e0127600. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Yen, Y. Characterization of the human ribonucleotide reductase M2 subunit gene; genomic structure and promoter analyses. Cytogenet. Cell Genet. 2001, 95, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.C.; Motzer, R.J.; Tong, Y.; Bosl, G.J. Computerized quantitation of synergism and antagonism of taxol, topotecan, and cisplatin against human teratocarcinoma cell growth: A rational approach to clinical protocol design. J. Natl. Cancer Inst. 1994, 86, 1517–1524. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.C.; Talalay, P. Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 1984, 22, 27–55. [Google Scholar] [CrossRef]
- Guo, J.R.; Chen, Q.Q.; Lam, C.W.; Zhang, W. Effects of karanjin on cell cycle arrest and apoptosis in human A549, HepG2 and HL-60 cancer cells. Biol. Res. 2015, 48, 1. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of Phyllanthus amarus Extract are available from the authors.



| Time (h) | PHA (µg/mL) | ||
|---|---|---|---|
| 25.0 | 50.0 | 100.0 | |
| 24 | 1.16 | 1.23 | 1.37 |
| 48 | 0.76 | 0.66 | 0.71 |
| MRM Transition | Control | F | P | P-F | |
|---|---|---|---|---|---|
| ATP | 506.0→159.0 | 10904.7 ± 275.1 | 18975.6 ± 1728.7 ** | 9775.0 ± 936.9 * | 16173.0 ± 1784.1 **,# |
| ADP | 426.0→328.0 | 2985.5 ± 89.5 | 49033 ± 207.5 ** | 2933.1 ± 190.5 | 4916.2 ± 709.2 ** |
| AMP | 346.0→79.3 | 469.5 ± 21.1 | 608.5 ± 49.7 ** | 444.1 ± 27.6 | 823.9 ± 156.21 **,## |
| CTP | 482.0→384.0 | 1601.3 ± 41.7 | 2860.0 ± 269.9 ** | 1523.9 ± 162.1 | 2116.5 ± 213.2 **,## |
| CDP | 402.0→159.0 | 430.3 ± 15.9 | 707.9 ± 35.9 ** | 468.4 ± 28.9 * | 697.7 ± 98.7 ** |
| CMP | 322.0→79.4 | 153.4 ± 7.01 | 200.2 ± 7.2 ** | 148.3 ± 8.6 | 232.2 ± 37.0 **,# |
| GTP | 522.0→423.9 | 5428.2 ± 110.7 | 8923.2 ± 807.2 ** | 4668.0 ± 547.8 ** | 7912.1 ± 807.8 **,# |
| GDP | 442.0→344.0 | 1263.4 ± 45.3 | 1935.3 ± 64.0 ** | 1203.3 ± 87.2 | 1858.9 ± 305.0 ** |
| GMP | 362.0→79.30 | 67.0 ± 2.8 | 81.3 ± 8.2 ** | 55.0 ± 4.1 ** | 115.8 ± 22.5 **,## |
| UTP | 482.9→385.0 | 4989.0 ± 114.2 | 8858.3 ± 774.9 ** | 5097.9 ± 517.1 | 7045.6 ± 741.2 **,## |
| UDP | 403.0→159.0 | 792.9 ± 38.2 | 1280.0 ± 69.9 ** | 844.9 ± 55.7 | 1261.1 ± 189.2 ** |
| UMP | 323.0→79.3 | 39.3 ± 1.8 | 49.0 ± 4.2 ** | 38.6 ± 2.3 | 56.4 ± 12.4 ** |
| MRM Transition | Control | F | P | P-F | |
|---|---|---|---|---|---|
| dATP | 490.0→159.1 | 30.34 ± 0.5 | 113.11 ± 10.2 ** | 32.45 ± 1.8 * | 126.9 ± 16.4 ** |
| dADP | 410.0→79.0 | 0.0568 ± 0.005 | 0.2789 ± 0.02 ** | 0.0526 ± 0.003 | 0.3414 ± 0.05 **,# |
| dAMP | 330.0→79.0 | 0.0068 ± 0.002 | 0.0354 ± 0.003 ** | 0.007 ± 0.001 | 0.0525 ± 0.01**,## |
| dCTP | 465.9→159.0 | 28.48 ± 0.6 | 138.3 ± 12.3 ** | 24.00 ± 1.8 ** | 126.6 ± 18.8 ** |
| dCDP | 386.0→79.0 | 0.0297 ± 0.005 | 0.1185 ± 0.01 ** | 0.0189 ± 0.002 ** | 0.1446 ± 0.03 ** |
| dCMP | 306.0→79.0 | 0.0499 ± 0.006 | 0.0394 ± 0.007 * | 0.0304 ± 0.008 ** | 0.0923 ± 0.016 **,## |
| dGTP | 581.0→152.1 | 40.35 ± 2.3 | 12.81 ± 0.50 ** | 34.22 ± 3.4 ** | 9.52 ± 0.84 **,## |
| dGDP | 501.0→152.0 | 2.12 ± 0.09 | 0.13 ± 0.07 ** | 2.22 ± 0.4 | 0.13 ± 0.06 ** |
| dGMP | 421.0→152.0 | 2.92 ± 0.1 | 0.28 ± 0.07 ** | 2.72 ± 0.4 | 0.35 ± 0.09 ** |
| dUTP | 467.0→159.0 | *UDL | *UDL | *UDL | *UDL |
| dUDP | 387.0→79.0 | 0.02 ± 0.01 | 0.22 ± 0.07 ** | 1.52 ± 3.4 | 0.59 ± 0.39 **,# |
| dUMP | 307.0→79.0 | 0.17 ± 0.03 | 76.38 ± 5.7 ** | 0.15 ± 0.05 | 137.79 ± 17.3 **,## |
| dTTP | 481.0→159.1 | 85.54 ± 1.0 | 40.61 ± 3.0 ** | 92.77 ± 7.2 * | 15.73 ± 1.8 **,## |
| dTDP | 401.0→79.0 | 2.66 ± 0.2 | 1.25 ± 0.05 ** | 3.11 ± 0.1 ** | 1.14 ± 0.07 **,## |
| dTMP | 321.0→79.0 | 0.0435 ± 0.005 | 0.004 ± 0.0008 ** | 0.049 ± 0.003 * | 0.0071 ± 0.004 ** |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, J.-R.; Chen, Q.-Q.; Lam, C.W.-K.; Wang, C.-Y.; Xu, F.-G.; Liu, B.-M.; Zhang, W. Effect of Phyllanthus amarus Extract on 5-Fluorouracil-Induced Perturbations in Ribonucleotide and Deoxyribonucleotide Pools in HepG2 Cell Line. Molecules 2016, 21, 1254. https://doi.org/10.3390/molecules21091254
Guo J-R, Chen Q-Q, Lam CW-K, Wang C-Y, Xu F-G, Liu B-M, Zhang W. Effect of Phyllanthus amarus Extract on 5-Fluorouracil-Induced Perturbations in Ribonucleotide and Deoxyribonucleotide Pools in HepG2 Cell Line. Molecules. 2016; 21(9):1254. https://doi.org/10.3390/molecules21091254
Chicago/Turabian StyleGuo, Jian-Ru, Qian-Qian Chen, Christopher Wai-Kei Lam, Cai-Yun Wang, Feng-Guo Xu, Bu-Ming Liu, and Wei Zhang. 2016. "Effect of Phyllanthus amarus Extract on 5-Fluorouracil-Induced Perturbations in Ribonucleotide and Deoxyribonucleotide Pools in HepG2 Cell Line" Molecules 21, no. 9: 1254. https://doi.org/10.3390/molecules21091254