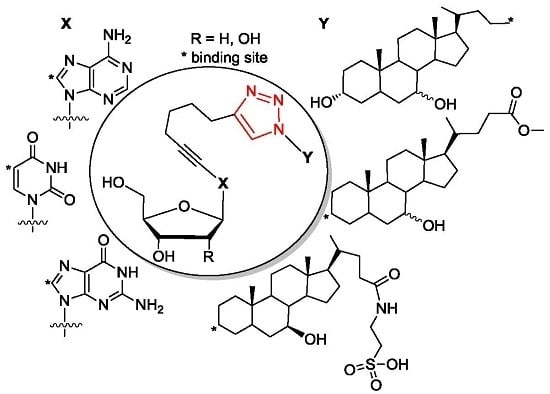
4.7.3. Method C (Purification of Conjugates with TUDCA-Azides)
The mixture was concentrated under reduced pressure, added with water and extracted with n-butanol. The organic layers was dried over Na2SO4, filtered and concentrated in vacuo. The crude white solid was washed twice with EtOH (10 mL) and dried with Et2O.
A-CDC. Colourless syrup, yield 80%; IR: ν (cm−1) 3418–3315 (O-H), 2928–2866 (C-H), 2241 (C≡C), 1693 (C=O), 1665–1524 (C=C,C=N); 1H-NMR: δ = 8.18 (br s, 1H), 7.88 (s, 1H), 7.58 (br s, 2H), 5.98 (d, J = 6.83 Hz 1H), 5.61–5.58 (m, 1H), 5.42 (d, J = 6.25 Hz 1H), 5.20 (d, J = 4.30 Hz, 1H), 5.12–4.98 (m, 1H), 4.32–4.16 (m, 3H), 3.98 (m, 1H), 3.72–3.42 (s, 7H), 2.74–2.59 (m, 5H), 2.38–2.16 (m, 3H), 2.01–0.95 (m, 25H), 0.90 (s, 3H), 0.86 (d, J = 6.44 Hz, 3H), 0.60 (s, 3H); 13C-NMR: δ = 173.8 (q), 156.0 (q), 153.2 (CH), 148.3 (q), 146.1 (q), 134.0 (q), 120.0 (q), 119.9 (CH), 97.4 (q), 89.3 (CH), 86.6 (CH), 71.5 (CH), 71.08 (CH), 70.3 (q), 66.1 (CH), 62.2 (CH2), 60.1 (CH), 55.4 (CH), 51.2 (CH3), 49.9 (CH), 41.9 (CH), 41.7 (q), 40.1 (CH2), 37.7 (CH2), 35.4 (CH2), 34.9 (q), 34.8 (CH), 34.4 (CH2), 32.2 (CH), 30.6 (CH2), 30.4 (CH2), 28.2 (CH2), 27.7 (CH2), 27.2 (CH2), 27.1 (CH2), 24.6 (CH2), 23.1 (CH2), 22.6 (CH3), 20.3 (CH2), 18.4 (CH2), 18.1 (CH3), 11.6 (CH3). HRMS calculated for [C43H62N8O7 + H]+ 803.4814, found 803.4819.
A-UDC. Colourless syrup, yield 78%; IR: ν (cm−1) 3411 (O-H), 2926–2865 (C–H), 2240 (C≡C), 1693 (C=O), 1660–1524 (C=C,C=N); 1H-NMR: δ = 8.18 (br s, 1H), 8.02 (s, 1H), 7.58 (br s, 2H), 5.98 (d, J = 6.83 Hz, 1H), 5.61–5.58 (m, 1H), 5.42 (d, J = 6.23 Hz, 1H), 5.20 (d, J = 4.31 Hz, 1H), 5.12–4.98 (m, 1H), 4.38 (br, s, 1H), 4.18 (br, s, 1H), 3.98 (br, s, 1H), 3.91 (br, s, 1H), 3.71–3.62 (m, 1H), 3.58–3.45 (m, 4H), 2.74–2.59 (m, 5H), 2.38–2.11 (m, 5H), 2.01–0.95 (m, 25H), 0.92 (s, 3H), 0.84 (d, J = 6.44 Hz, 3H), 0.60 (s, 3H); 13C-NMR: δ = 174.2 (q), 156.4 (q), 153.4 (CH), 148.7 (q), 142.8 (q), 133.9 (q), 120.4 (q), 120.2 (CH), 98.0 (q), 88.8 (CH), 85.6 (CH), 71.8 (q), 70.8 (CH), 66.5 (CH), 62.7 (CH2), 60.5 (CH), 55.9 (CH), 51.6 (CH3), 50.4 (CH), 42.4 (q), 42.1 (CH), 39.6 (CH), 38.0 (CH2), 37.7 (CH2), 35.9 (CH2), 35.3 (CH), 35.3 (CH2), 34.9 (q), 32.7 (CH2), 31.1 (CH), 30.8 (CH2), 28.7 (CH2), 28.2 (CH2), 27.6 (CH2), 27.5 (CH2), 25.0 (CH2), 23.5 (CH2), 23.1 (CH2), 20.7 (CH3), 18.8 (CH2), 18.6 (CH3), 12.1 (CH3); HRMS calculated for [C43H62N8O7 + H]+ 803.4814, found 803.4829.
dG-CDC. Colourless syrup, yield 76%; IR: ν (cm−1) 3327 (O-H), 2933–2865 (C-H), 2243 (C≡C), 1692 (C=O), 1629–1568 (C=C,C=N); 1H-NMR: δ = 10.82 (br, s, 1H), 7.85 (s, 1H), 6.57 (br, s, 2H), 6.25–6.19 (m, 1H), 5.22 (br, s, 1H), 4.86 (br, s, 1H), 4.4–4.16 (m, 3H), 3.8 (m, 1H), 3.6–3.58 (m, 2H), 3.59 (s, 3H), 3.57–3.42 (m, 2H), 3.15–2.99 (m, 1H), 2.71–2.62 (m, 2H), 2.53 (m, 2H), 2.38–2.01 (m, 5H), 1.92–0.99 (m, 25H), 0.89 (s, 3H), 0.86 (d, J = 6.40 Hz, 3H), 0.60 (s, 3H); 13C-NMR: δ = 173.6 (q), 156.5 (q), 153.6 (q), 150.5 (q), 146.3 (q), 130.1 (q), 121.6 (CH), 116.8 (q), 95.2 (q), 87.6 (CH), 84.6 (CH), 70.9 (q), 70.6 (CH), 70.5 (CH), 66.0 (CH), 62.0 (CH2), 60.0 (CH), 55.4 (CH), 51.1 (CH3), 49.8 (CH), 41.9 (CH), 41.6 (q), 40.0 (CH2), 39.6 (CH), 38.8 (CH2), 37.1 (CH2), 35.4 (CH2), 34.7 (q), 34.3 (CH2), 32.1 (CH), 30.6 (CH2), 30.3 (CH2), 28.1 (CH2), 27.7 (CH2), 27.1 (CH2), 24.5 (CH2), 23.0 (CH2), 22.6 (CH3), 20.2 (CH2), 18.3 (CH2), 18.2 (CH3), 11.6 (CH3). HRMS calculated for [C43H62N8O7 + H]+ 803.4814, found 803.4815.
dG-UDC. Colourless syrup, yield 78%; IR: ν (cm−1) 3312 (O-H), 2929–2860 (C-H), 2240 (C≡C), 1686 (C=O), 1601–1565 (C=C,C=N); 1H-NMR: δ = 10.78 (br, s, 1H), 7.99 (s, 1H), 6.48 (br, s, 2H), 6.28–6.18 (m, 1H), 5.22 (br, s, 1H), 4.93–4.82 (m, 1H), 4,38 (br, s, 2H), 3.91 (d, J = 6.45 Hz, 1H), 3.79–3.75 (m, 1H), 3.62–3.41 (m, 5H), 3.19–2.98 (m, 1H), 2.68–0.99 (m, 36H), 0.97 (s, 3H), 0.86 (d, J = 6.44 Hz, 3H), 0.59 (s, 3H); 13C-NMR: δ = 173.7 (q), 158.3 (q), 155.9 (q), 153.6 (q), 150.5 (q), 149.9 (q), 129.5 (q), 119.8 (CH), 94.0 (q), 87.6 (CH), 83.6 (CH), 71.8 (q), 71.0 (CH), 68.9 (CH), 68.0 (CH), 62.0 (CH2), 59.5 (CH), 55.2 (CH), 54.5 (CH), 51.1 (CH3), 42.9 (CH), 42.4 (CH), 41.8 (q), 40.0 (CH2), 37.2 (CH2), 36.8 (CH2), 34.9 (CH2), 34.7 (CH), 34.0 (q), 33.7 (CH2), 30.6 (CH2), 30.2 (CH2), 28.1 (CH2), 27.5 (CH2), 27.1 (CH2), 26.5 (CH2), 24.5 (CH2), 23.8 (CH2), 23.1 (CH3), 20.8 (CH2), 18.2 (CH2), 18.1 (CH3), 11.9 (CH3). HRMS calculated for [C43H62N8O7 + H]+ 803.4814, found 803.4816.
G-CDC. Colourless syrup, yield 80%; IR: ν (cm−1) 3330 (O-H), 2918–2850 (C-H), 2238 (C≡C), 1736 (C=O), 1645–1580 (C=C,C=N); 1H-NMR: δ = 10.78 (br, s, 1H), 7.88 (s, 1H), 6.48 (br, s, 2H), 5.78 (d, J = 6.44 Hz, 1H), 5.40 (d, J = 6.25 Hz, 1H), 5.11 (d, J = 4.88, 1H), 4.99–4.82 (m, 2H), 4,32–4.18 (m, 2H), 4.15 (br, s, 1H), 3.82 (br, s, 1H), 3.63–3.41 (m, 4H), 2.75–2.15 (m, 5H), 1.96–0.99 (m, 31 H), 0.88 (s, 3H), 0.84 (d, J = 6.44 Hz, 3H), 0.60 (s, 3H); 13C-NMR: δ = 173.7 (q), 157.4 (q), 155.9 (q), 153.7 (q), 150.7 (q), 130.2 (q), 119.9 (CH), 116.8 (q), 95.2 (q), 88.3 (CH), 85.5 (CH), 70.9 (q), 70.6 (CH), 70.5 (CH), 66.0 (CH), 62.0 (CH2), 60.0 (CH), 55.4 (CH), 51.1 (CH3), 49.8 (CH), 41.9 (CH), 41.6 (q), 40.0 (CH2), 39.6 (CH), 38.8 (CH2), 37.1 (CH2), 35.4 (CH2), 34.8 (CH), 34.7 (q), 34.3 (CH2), 32.1 (CH), 30.6 (CH2), 30.3 (CH2), 28.1 (CH2), 27.7 (CH2), 27.1 (CH2), 24.5 (CH2), 23.0 (CH2), 22.5 (CH3), 20.2 (CH2), 18.3 (CH2), 18.1 (CH3), 11.6 (CH3). HRMS calculated for [C43H62N8O8 + H]+ 819.4763, found 819.4768.
G-UDC. Light yellow syrup, yield 78%; IR: ν (cm−1) 3327 (O-H), 2930–2875 (C-H), 2238 (C≡C), 1735 (C=O), 1645–1572 (C=C,C=N); 1H-NMR: δ = 10.78 (br, s, 1H), 8.02 (s, 1H), 6.48 (br, s, 2H), 5.78 (d, J = 6.43 Hz, 1H), 5.40 (d, J = 6.24 Hz, 1H), 5.11 (d, J = 4.88, 1H), 4.99–4.82 (m, 2H), 4.42–4.35(m, 1H), 4,18–4.12 (m, 1H), 3.91 (d, J = 6.64 Hz, 1H), 3.823.78 (m, 1H), 3.68–3.38 (m, 6H), 2.71–0.98 (m, 34H), 0.92 (s, 3H), 0.83 (d, J = 6.44 Hz, 3H), 0.58 (s, 3H); 13C-NMR: δ = 173.5 (q), 156.0 (q), 153.7 (q), 150.7 (q), 146.3 (q), 130.2 (q), 121.6 (CH), 116.8 (q), 95.2 (q), 88.3 (CH), 85.5 (CH), 71.8 (q), 71.0 (CH), 68.9 (CH), 68.0 (CH), 62.0 (CH2), 59.5 (CH), 55.2 (CH), 54.5 (CH), 51.1 (CH3), 42.9 (CH), 42.4 (CH), 41.8 (q), 40.0 (CH2), 37.2 (CH2), 36.8 (CH2), 34.9 (CH2), 34.7 (CH), 34.0 (q), 33.7 (CH2), 30.6 (CH2), 30.2 (CH2), 28.1 (CH2), 27.5 (CH2), 27.1 (CH2), 26.5 (CH2), 24.5 (CH2), 23.8 (CH2), 23.1 (CH3), 20.8 (CH2), 18.2 (CH2), 18.1 (CH3), 11.9 (CH3). HRMS calculated for [C43H62N8O7 + H]+ 819.4763, found 819.4766.
dU-CDC. Amorphous white solid, yield 90%; IR: ν (cm−1) 3440–3310 (O-H), 2930–2861 (C-H), 2244 (C≡C), 1693 (C=O), 1633–1565 (C=C,C=N); 1H-NMR: δ = 8.62 (s, 1H), 7.81 (s, 1H), 6.42 (s, 1H), 6.17–6.12 (m, 1H), 5.28 (d, J = 4.3 Hz, 1H), 5.14–5.09 (m, 1H), 4.24–4.18 (m, 3H), 3.88 (q, J = 3.71 Hz, 1H), 3.67–3.58 (m, 3H), 3.57 (s, 3H), 2.78–2.58 (m, 5H), 2.40–2.24 (m, 2H), 2.23–2.13 (m, 1H), 2.08–1.98 (m, 1H), 1.92–0.95 (m, 27H), 0.89 (s, 3H), 0.86 (d, J = 6.44 Hz, 3H), 0.60 (s, 3H); 13C-NMR: δ = 173.8 (q), 171.2 (q), 158.1 (q), 153.8 (q), 146.1 (q), 136.8 (CH), 119.8 (CH), 106.4 (q) , 99.9 (q), 88.1 (CH), 87.4 (CH), 69.7 (CH), 66.1 (CH), 60.8 (CH), 60.1 (CH2), 55.5 (CH), 51.2 (CH), 49.9 (CH3), 42.0 (q), 41.7 (CH), 41.21 (CH), 37.3 (CH2), 35.4 (CH2), 34.9 (CH), 34.8 (CH2), 34.4 (q), 32.2 (CH), 30.7 (CH2), 30.4 (CH2), 28.3 (CH2), 27.8 (CH2), 27.2 (CH2), 27.1 (CH2), 26.0 (CH2), 24.7 (CH2), 23.1 (CH2), 22.6 (CH3), 20.3 (CH2), 18.2 (CH3), 11.7 (CH3); HRMS calculated for [C42H61N5O8 + H]+ 764.4592, found 764.4602.
dU-UDC. Amorphous white solid, yield 70%; IR: ν (cm−1) 3447–3309 (O-H), 3016–2942 (C-H), 2254 (C≡C), 1690 (C=O), 1645–1522 (C=C,C=N); 1H-NMR: δ = 11.58 (s, 1H), 8.18 (s, 1H), 7.98 (s, 1H), 6.12–6.07 (m, 1H), 5.28–5.21 (m, 1H), 5.18–5.15 (m, 1H), 4.42–4.28 (m, 1H), 4.21 (br s, 1H), 3.95 (d, J = 6.64 Hz, 1H), 3.78 (br s, 1H), 3.65–3.48 (m, 4H), 2.65–2.58 (m, 2H), 2.41–0.98 (m, 36H), 0.95 (s, 3H), 0.85 (d, J = 6.44 Hz, 3H), 0.62 (s, 3H); 13C-NMR: δ = 175.4 (q), 171.8 (q), 162.2 (q), 149.9 (q), 146.7 (q), 143.2 (CH), 120.3 (CH), 99.4 (q), 88.0 (CH), 85.0 (CH), 80.7 (CH), 70.6 (CH), 69.5 (CH), 61.4 (CH2), 60.0 (CH), 55.8 (CH), 51.6 (CH3), 43.5 (CH), 43.0 (CH), 42.5 (q), 40.5 (CH2), 40.0 (CH2), 38.8 (CH), 37.74 (CH2), 35.4 (CH2), 35.2 (CH), 34.6 (CH2), 34.3 (q), 31.2 (CH2), 30.9 (CH2), 29.5 (CH2), 28.6 (CH2), 28.2 (CH2), 27.1 (CH2), 25.1 (CH2), 23.6 (CH3), 21.4 (CH2), 19.0 (CH2), 18.7 (CH3), 12.4 (CH3); HRMS calculated for [C42H61N5O8 + H]+ 764.4592, found 764.4600.
dA-nor-CDC. Light yellow syrup, yield 68%; IR: ν (cm−1) 3440–3332 (O-H), 2926–2865 (C-H), 2242 (C≡C), 1650–1570 (C=C,C=N); 1H-NMR: δ = 8.16 (br, s, 1H), 7.89 (s, 1H), 7.58 (br, s, 2H), 6.42–6.38 (m, 1H), 5.43–5.39 (m, 1H), 5.32 (d, J = 4.30 Hz, 1H), 4.45 (br, s, 1H), 4.36–4.22 (m, 3H), 4.08 (d, J = 4.12 Hz, 1H), 3.90–3.85 (m, 1H), 3.72–3.42 (m, 3H), 3.20–3.03 (m, 2H), 2.71–2.57 (m, 4H), 2.19–2.09 (m, 2H), 1.97–0.99 (m, 27 H), 0.91 (d, J = 6.25 Hz, 3H), 0.75 (s, 3H), 0.48 (s, 3H); 13C-NMR: δ = 155.8 (q), 152.9 (CH), 148.2 (q), 146.3 (q), 133.3 (q), 121.6 (CH), 119.0 (q), 97.4 (q), 88.2 (CH), 85.1 (CH), 71.3 (CH), 70.2 (CH), 69.2 (q), 66.0 (CH), 62.1 (CH2), 55.2 (CH), 50.1 (CH2), 49.8 (CH), 46.8 (CH2), 41.8 (q), 41.3 (CH), 40.3 (CH), 39.2 (CH2), 37.5 (CH2), 36.0 (CH2), 35.2 (CH2), 34.6 (q), 34.5 (CH2), 32.9 (CH), 32.1 (CH), 30.4 (CH2), 28.0 (CH2), 27.7 (CH2), 26.7 (CH2), 24.2 (CH2), 22.9 (CH2), 22.5 (CH3), 20.0 (CH2), 18.1 (CH2), 18.1 (CH3), 11.3 (CH3); HRMS calculated for [C41H60N8O5 + H]+ 745.4759, found 745.4767.
dA-nor-UDC. Light yellow syrup, 72%; IR: ν (cm−1) 3440–3317 (O-H), 2928–2861 (C-H), 2243 (C≡C), 1603–1569 (C=C,C=N); 1H-NMR: δ = 8.18 (br, s, 1H), 7.88 (s, 1H), 7.55 (br, s, 2H), 6.42–6.39 (m, 1H), 5.42–5.39 (m, 1H), 5.37 (d, J = 4.30 Hz, 1H), 4.43 (br, s, 2H), 4.38–4.20 (m, 2H), 3.92–3.81 (m, 2H), 3.70–3.61 (m, 1H), 3.53–3.42 (m, 1H), 3.38–3.18 (m, 3H), 3.17–3.04 (m, 1H), 2.72–2.57 (m, 4H), 2.20–2.11 (m, 1H), 1.97–0.99 (m, 27 H), 0.91 (d, J = 6.25 Hz, 3H), 0.81 (s, 3H), 0.52 (s, 3H); 13C-NMR: δ = 156.4 (q), 153.3 (CH), 148.7 (q), 147.0 (q), 133.9 (q), 122.3 (CH), 120.0 (q), 97.9 (q), 88.8 (CH), 85.7 (CH), 71.8 (CH), 70.8 (q) 70.2 (CH), 69.8 (CH), 62.7 (CH2), 56.2 (CH), 54.9 (CH), 48.8 (CH2), 47.5 (CH2), 43.5 (q), 43.4 (CH), 42.6 (CH), 40.0 (CH2), 39.1 (CH), 38.1 (CH2), 37.7 (CH2), 36.7 (CH2), 35.2 (CH2), 34.7 (CH2), 34.1 (q), 33.4 (CH), 30.7 (CH2), 28.6 (CH2), 27.3 (CH2), 27.1 (CH2), 24.8 (CH2), 23.7 (CH3), 21.2 (CH2), 18.9 (CH3), 18.8 (CH2), 12.3 (CH3); HRMS calculated for [C41H60N8O5 + H]+ 745.4759, found 745.4765.
A-nor-CDC. Amorphous white solid, yield 68%; IR: ν (cm−1) 3443–3318 (O-H), 2932–2863 (C-H), 2241 (C≡C), 1640–1570 (C=C,C=N); 1H-NMR: δ = 8.12 (s, 1H), 7.87 (s, 1H), 7.58 (br, s, 2H), 5.92 (d, J = 6.83 Hz, 1H), 5.60–5.55 (m, 1H), 5.41 (d, J = 6.25 Hz, 1H), 5.20 (d, J = 4.30 Hz, 1H), 5.00–4.96 (m, 1H), 4.32–4.28 (m, 2H), 4.19–4.12 (m, 1H), 4.08 (d, J = 4.12 Hz, 1H), 3.98–3.96 (m, 1H), 3.72–3.44 (m, 3H), 3.20–3.12 (m, 1H), 2.68–2.57 (m, 4H), 2.38–2.16 (m, 3H), 1.98–0.99 (m, 26H), 0.95 (d, J = 6.25 Hz, 3H), 0.79 (s, 3H), 0.52 (s, 3H); 13C-NMR: δ = 155.8 (q), 152.9 (CH), 148.1 (q), 146.3 (q), 133.9 (q), 121.6 (CH), 119.0 (q), 97.2 (q), 89.2 (CH), 86.5 (CH), 71.4 (CH), 70.9 (CH), 70.2 (CH), 69.5 (q), 66.0 (CH), 62.1 (CH2), 55.2 (CH), 49.9 (CH), 46.8 (CH2), 41.8 (q), 41.2 (CH), 40.0 (CH), 36.1 (CH2), 35.2 (CH2), 34.4 (q), 34.6 (CH2), 33.0 (CH), 32.1 (CH), 30.4 (CH2), 28.9 (CH2), 28.6 (CH2), 28.0 (CH2), 27.7 (CH2), 26.8 (CH2), 24.3 (CH2), 23.0 (CH2), 22.6 (CH3), 20.1 (CH2), 18.2 (CH2), 18.2 (CH3), 11.4 (CH3); HRMS calculated for [C41H60N8O6 + H]+ 761.4708, found 761.4704.
A-nor-UDC. Amorphous white solid, yield 71%; IR: ν (cm−1) 3440–3318 (O-H), 2930–2863 (C-H), 2247 (C≡C), 1640–1570 (C=C,C=N); 1H-NMR: δ = 8.20 (br, s, 1H), 7.89 (s, 1H), 7.58 (br, s, 2H), 5.92 (d, J = 6.83 Hz, 1H), 5.68–5.59 (m, 1H), 5.41 (d, J = 6.25 Hz, 1H), 5.22 (d, J = 4.30 Hz, 1H), 5.00–4.96 (m, 1H), 4.52 (d, J = 4.30 Hz, 1H), 4.35–4.12 (m, 3H), 3.98–3.88 (m, 3H), 3.80–3.44 (m, 3H), 2.71–2.52 (m, 4H), 1.97–0.99 (m, 28H), 0.93 (d, J = 6.25 Hz, 3H), 0.82 (s, 3H), 0.53 (s, 3H); 13C-NMR: δ = 156.4 (q), 153.5 (CH), 148.7 (q), 146.8 (q), 134.4 (q), 122.3 (CH), 119.5 (q), 97.8 (q), 89.7 (CH), 87.0 (CH), 72.0 (CH), 71.4 (CH), 70.7 (q), 70.1 (CH), 69.9 (CH), 62.6 (CH2), 56.2 (CH), 54.9 (CH), 47.4 (CH2), 43.5 (q), 43.4 (CH), 42.6 (CH), 39.1 (CH), 38.1 (CH2), 37.7 (CH2), 36.7 (CH2), 35.3 (CH2), 34.2 (q), 33.4 (CH), 30.6 (CH2), 28.6 (CH2), 27.3 (CH2), 27.1 (CH2), 24.8 (CH2), 23.7 (CH3), 21.2 (CH2), 18.9 (CH3), 18.8 (CH2), 18.3 (CH2), 12.3 (CH3); HRMS calculated for [C41H60N8O6 + H]+ 761.4708, found 761.4715.
dG-nor-CDC. Amorphous white solid, yield 64%; IR: ν (cm−1) 3402–3320 (O-H), 2924–2850 (C-H), 2233 (C≡C), 1688 (C=O), 1609–1523 (C=C,C=N); 1H-NMR: δ = 10.78 (br, s, 1H), 7.88 (s, 1H), 6.49 (br, s, 2H), 6.24–6.18 (m, 1H), 5.23 (br, s, 1H), 4.91 (br, s, 1H), 4.42–4.21 (m, 4H), 4.08 (br, s, 1H), 3.82–3.75 (m, 1H), 3.64–3.41 (m, 3H), 3.11–2.98 (m, 2H), 2.69–2.59 (m, 2H), 2.19–0.98 (m, 31H), 0.98 (d, J = 6.25 Hz, 3H), 0.78 (s, 3H), 0.52 (s, 3H); 13C-NMR: δ = 156.0 (q), 153.6 (q), 150.4 (q), 146.3 (q), 129.6 (q), 121.6 (CH), 116.8 (q), 95.1 (q), 87.6 (CH), 83.6 (CH), 71.4 (CH), 70.2 (CH), 69.9 (q), 66.0 (CH), 62.0 (CH2), 55.2 (CH), 49.8 (CH), 46.8 (CH2), 41.8 (q), 41.3 (CH), 40.3 (CH), 39.6 (CH2), 39.4 (CH), 38.9 (CH2), 36.8 (CH2), 36.0 (CH2), 35.2 (CH2), 34.6 (CH2), 32.9 (q), 32.1 (CH), 30.4 (CH2), 28.0 (CH2), 27.7 (CH2), 26.8 (CH2), 24.2 (CH2), 22.9 (CH2), 22.6 (CH3), 20.1 (CH2), 18.2 (CH2), 18.1 (CH3), 11.4 (CH3). HRMS calculated for [C41H60N8O6 + H]+ 761.4708, found 761.4705.
dG-nor-UDC. Amorphous white solid, yield 65%; IR: ν (cm−1) 3405–3322 (O-H), 2932–2852 (C-H), 2243 (C≡C), 1687 (C=O), 1645–1523 (C=C,C=N); 1H-NMR: δ = 7.87 (s, 1H), 6.48 (br, s, 2H), 6.25–6.19 (m, 1H), 5.22 (br, s, 1H), 4.89 (br, s, 1H), 4,48–4.22 (m, 4H), 3.90–3.75 (m, 3H), 3.62–3.41 (m, 2H), 3.09–2.98 (m, 1H), 2.75–2.62 (m, 2H), 2.32–0.99 (m, 33H), 0.98 (d, J = 6.25 Hz, 3H), 0.88 (s, 3H), 0.57 (s, 3H); 13C-NMR: δ = 155.9 (q), 153.5 (q), 150.4 (q), 146.3 (q), 129.7 (q), 121.7 (CH), 116.7 (q), 95.2 (q), 87.6 (CH), 83.6 (CH), 71.0 (CH), 69.9 (q), 69.6 (CH), 69.3 (CH), 62.0 (CH2), 55.7 (CH), 54.3 (CH), 46.9 (CH2), 42.9 (q), 42.8 (CH), 42.0 (CH), 39.5 (CH2), 38.7 (CH), 37.6 (CH2), 37.1 (CH2), 36.8 (CH2), 36.1 (CH2), 34.7 (CH2), 33.6 (q), 32.8 (CH), 30.1 (CH2), 28.9 (CH2), 28.0 (CH2), 26.8 (CH2), 26.5 (CH2), 24.2 (CH2), 23.2 (CH3), 20.7 (CH2), 18.3 (CH3), 18.2 (CH2), 11.8 (CH3). HRMS calculated for [C41H60N8O6 + H]+ 761.4708, found 761.4705.
G-nor-CDC. Light yellow syrup, yield 69%; IR: ν (cm−1) 3409–3310 (O-H), 2926–2857 (C-H), 2243 (C≡C), 1693 (C=O), 1640–1526 (C=C,C=N); 1H-NMR: δ = 10.87 (br, s, 1H), 7.89 (s, 1H), 6.52 (br, s, 2H), 5.78 (d, J = 6.44 Hz, 1H), 5.40 (d, J = 6.25 Hz, 1H), 5,05 (br, s, 1H), 4.99–4.84 (m, 2H), 4.38–4.21 (m, 4H), 4.09 (br, s, 2H), 3.83 (br, s, 1H), 3.68–3.55 (m, 2H), 3.54–3.43 (m, 1H), 3.21–3.09 (m, 2H), 2.70–2.60 (m, 2H), 2.55–2.48 (m, 2H), 2.22–2.15 (m, 2H), 1.98–1.02 (m, 23H), 0.93 (d, J = 6.25 Hz, 3H), 0.79 (s, 3H), 0.52 (s, 3H); 13C-NMR: δ = 156.0 (q), 153.6 (q), 150.7 (q), 146.4 (q), 130.3 (q), 121.6 (CH), 116.9 (q), 95.2 (q), 88.4 (CH), 85.5 (CH), 72.0 (CH), 70.2 (CH), 69.8 (q), 66.0 (CH), 62.1 (CH2), 55.5 (CH), 49.9 (CH), 48.2 (CH2), 41.9 (CH), 41.7 (q), 41.3 (CH), 41.0 (CH), 39.6 (CH2), 36.1 (CH), 35.2 (CH2), 34.6 (CH2), 34.1 (CH2), 33.1 (q), 32.1 (CH), 30.4 (CH2), 28.0 (CH2), 27.8 (CH2), 27.0 (CH2), 26.5 (CH2), 24.3 (CH2), 23.0 (CH2), 22.6 (CH3), 20.1 (CH2), 18.1 (CH3), 18.0 (CH2), 11.5 (CH3); HRMS calculated for [C41H60N8O7 + H]+ 777.4657, found 777.4657.
G-nor-UDC. Light yellow syrup, yield 68%; IR: ν (cm−1) 3413–3314 (O-H), 2930–2860 (C-H), 2238 (C≡C), 1693 (C=O), 1640–1526 (C=C,C=N); 1H-NMR: δ = 10.79 (br, s, 1H), 7.88 (s, 1H), 6.50 (br, s, 2H), 5.78 (d, J = 6.44 Hz, 1H), 5.40 (d, J = 6.25 Hz, 1H), 5,05 (br, s, 1H), 4.99–4.83 (m, 2H), 4.42 (br, s, 1H), 4.39–4.18 (m, 2H), 4.04 (br, s, 1H), 3.98–3.78 (m, 2H), 3.72–3.58 (m, 1H), 3.57–3.42 (m, 1H), 2.78–2.45 (m, 4H), 1.98–0.99 (m, 30H), 0.98 (d, J = 6.25 Hz, 3H), 0.82 (s, 3H), 0.58 (s, 3H); 13C-NMR: δ = 156.0 (q), 153.8 (q), 150.7 (q), 146.4 (q), 130.3 (q), 121.8 (CH), 116.9 (q), 95.1 (q), 88.4 (CH), 85.6 (CH), 71.0 (q), 70.7 (CH), 70.6 (CH), 69.7 (CH), 69.4 (CH), 62.1 (CH2), 55.8 (CH), 54.5 (CH), 47.0 (CH2), 43.1 (q), 43.0 (CH), 42.2 (CH), 40.0 (CH2), 38.7 (CH), 37.7 (CH2), 37.3 (CH2), 36.2 (CH2), 34.8 (CH2), 33.7 (q), 33.0 (CH), 30.2 (CH2), 28.2 (CH2), 27.0 (CH2), 26.7 (CH2), 24.4 (CH2), 23.3 (CH3), 20.8 (CH2), 18.4 (CH3), 18.3 (CH2), 11.9 (CH3); HRMS calculated for [C41H60N8O7 + H]+ 776.4658, found 776.4668.
dU-nor-CDC. Light yellow syrup, yield 71%; IR: ν (cm−1) 3446 (O-H), 3100–2850 (C-H), 2255 (C≡C), 1665 (C=O), 1634–1565 (C=C,C=N); 1H-NMR: δ = 8.65 (s, 1H), 7.82 (s, 1H), 6.39 (s, 1H), 6.19–6.09 (m, 1H), 5.23 (br, s, 1H), 5.10 (br, s, 1H), 4.38–4.17 (m, 3H), 4.05 (br, s, 1H), 3.86 (br, s, 1H), 3.69–3.52 (m, 3H), 3.21–3.04 (m, 1H), 2.71–2.58 (m, 3H), 2.39–2.25 (m, 1H), 2.21–2.03 (m, 1H), 2.03–0.99 (m, 30 H), 0.93 (d, J = 6.25 Hz, 3H), 0.79 (s, 3H), 0.44 (s, 3H); 13C-NMR: δ = 158.0 (q), 153.6 (q), 146.3 (q), 136.4 (CH), 121.6 (CH), 106.2 (q), 99.7 (q), 88.0 (CH), 87.3 (CH), 70.2 (CH), 69.6 (CH), 69.5 (q), 66.0 (CH), 60.7 (CH2), 55.2 (CH), 49.9 (CH), 46.8 (CH2), 41.8 (q), 41.3 (CH), 41.1 (CH2), 40.3 (CH), 39.2 (CH2), 39.0 (CH2), 36.0 (CH2), 35.2 (CH2), 34.7 (q), 34.6 (CH2), 32.9 (CH), 32.1 (CH), 30.4 (CH2), 28.1 (CH2), 27.7 (CH2), 27.0 (CH2), 25.6 (CH2), 24.4 (CH2), 22.9 (CH2), 22.6 (CH3), 20.1 (CH2), 18.1 (CH3), 11.3 (CH3); HRMS calculated for [C40H59N5O7 + H]+ 722.4487, found 722.4492.
dU-nor-UDC. Light yellow syrup, yield 70%; IR: ν (cm−1) 3441 (O-H), 3016–2862 (C-H), 2255 (C≡C), 1691 (C=O), 1640–1572 (C=C,C=N); 1H-NMR: δ = 11.52 (s, 1H), 8.12 (s, 1H), 7.85 (s, 1H), 6.12–6.09 (m, 1H), 5.23 (br, s, 1H), 5.08 (br, s, 1H), 4.42 (br, s, 1H), 4.38–4.18 (m, 3H), 3.85 (br, s, 1H), 3.79 (br, s, 1H), 3.63–3.51 (m, 2H), 3.30–3.19 (m, 2H), 2.78–2.55 (m, 2H), 2.40–2.31 (m, 2H), 2.12–2.02 (m, 2H), 1.98–0.99 (m, 28 H), 0.92 (d, J = 6.25 Hz, 3H), 0.83 (s, 3H), 0.55 (s, 3H); 13C-NMR: δ = 161.9 (q), 149.3 (q), 146.6 (q), 142.6 (CH), 121.6 (CH), 98.9 (q), 92.9 (q), 87.4 (CH), 84.4 (CH), 72.9 (q), 70.0 (CH), 69.6 (CH), 69.3 (CH), 60.8 (CH2), 55.7 (CH), 54.4 (CH), 48.2 (CH2), 46.9 (CH2), 43.0 (q), 42.9 (CH), 42.0 (CH), 37.6 (CH2), 37.1 (CH2), 36.1 (CH2), 34.7 (CH2), 34.2 (CH2), 33.6 (q), 33.0 (CH), 32.9 (CH), 30.1 (CH2), 28.1 (CH2), 28.0 (CH2), 27.4 (CH2), 26.5 (CH2), 24.3 (CH2), 23.2 (CH3), 20.7 (CH2), 18.4 (CH2), 18.3 (CH3), 11.8 (CH3); HRMS calculated for [C40H59N5O7 + H]+ 722.4487, found 722.4490.
dA-TUDCA. Light yellow syrup, yield 68%; IR: ν (cm−1) 3340–3284 (O-H), 2918–2860 (C-H), 2240 (C≡C), 1651 (C=O), 1640–1514 (C=C,C=N); 1H-NMR: δ = 8.16 (s, 1H), 8.02 (s, 1H), 7.70–7.61 (m, 1H), 7.52 (br, s, 2H), 6.43–6.38 (m, 1H), 5.42–5.25 (m, 2H), 4.48–4.12 (m, 5H), 3.95–3.82 (m, 2H), 3.72–3.59 (m, 1H), 3.55–3.41 (m, 1H), 3.40–3.21 (m, 5H), 3.19–3.12 (m, 2H), 2.85–2.71 (m, 2H), 2.69–2.58 (m, 3H), 2.28–0.99 (m, 27H), 0.93 (s, 3H), 0.88 (d, J = 6.41 Hz, 3H), 0.60 (s, 3H); 13C-NMR: δ = 155.8 (q), 152.9 (q), 148.2 (q), 146.3 (q), 133.3 (q), 121.7 (CH), 119.2 (CH), 97.4 (q), 88.2 (CH), 85.1 (CH), 72.0 (q), 71.2 (CH), 70.2 (q), 69.6 (CH), 69.3 (CH), 62.1 (CH2), 55.6 (CH), 54.3 (CH), 50.1 (CH2), 46.9 (CH2), 42.9 (q), 42.8 (CH), 42.0 (CH), 40.3 (CH), 39.9 (CH2), 39.3 (CH2), 38.6 (CH), 37.6 (CH2), 37.5 (CH2), 37.1 (CH2), 36.1 (CH2), 34.7 (CH2), 33.6 (q), 32.9 (CH2), 30.1 (CH2), 28.1 (CH2), 26.7 (CH2), 26.5 (CH2), 24.2 (CH2), 23.2 (CH3), 20.7 (CH2), 18.3 (CH3), 18.2 (CH2), 11.8 (CH3); HRMS calculated for [C44H65N9O8S + H]+ 880.4749, found 880.4751.
A-TUDCA. White yellow syrup, yield 63%; IR: ν (cm−1) 3340–3311 (O-H), 2929–2870 (C-H), 2243 (C≡C), 1640 (C=O), 1650–1527 (C=C,C=N); 1H-NMR: δ = 8.16 (s, 1H), 8.02 (s, 1H), 7.73–7.63 (m, 1H), 7.58 (br, s, 2H), 5.98 (d, J = 6.83 Hz, 1H), 5.60–5.55 (m, 1H), 5.43 (d, J = 6.25 Hz, 1H), 5.22 (d, J = 4.58 Hz, 1H), 5.02–4.97 (m, 1H), 4.48–4.31 (m, 1H), 4.21–4.12 (m, 1H), 3.98–3.87 (m, 2H), 3.72–3.61 (m, 1H), 3.58–3.45(m, 1H), 3.33–3.23 (m, 3H), 2.69–2.55 (m, 3H), 2.15–0.99 (m, 35H), 0.92 (s, 3H), 0.88 (d, J = 6.41 Hz, 3H), 0.60 (s, 3H); 13C-NMR: δ = 172.6 (q), 156.4 (q), 153.4 (CH), 148.7 (q), 146.6 (q), 134.4 (q), 120.1 (CH), 97.8 (q), 89.7 (CH), 87.0 (CH), 72.0 (q), 71.9 (CH), 71.5 (CH), 70.7 (q), 69.5 (CH), 62.7 (CH2), 60.0 (CH), 55.8 (CH), 55.2 (CH), 51.0 (CH2), 43.5 (CH), 43.0 (CH), 42.0 (q), 40.0 (CH2), 38.8 (CH), 37.7 (CH2), 35.9 (CH2), 35.6 (CH2), 35.4 (CH), 34.6 (CH2), 34.2 (q), 33.0 (CH2), 32.0 (CH2), 28.6 (CH2), 28.1 (CH2), 27.5 (CH2), 27.1 (CH2), 25.1 (CH2), 23.6 (CH3), 21.4 (CH2), 18.9 (CH3), 18.8 (CH2), 18.3 (CH2), 12.5 (CH3); HRMS calculated for [C44H65N9O9S + H]+ 896.4698, found 896.4701.
dG-TUDCA. Yield 65%; IR: ν (cm−1) 3397 (O-H), 2930–2863 (C-H), 2254 (C≡C), 1690, 1646 (C=O), 1638–1552 (C=C,C=N); 1H-NMR: δ = 11.18 (s, 1H), 8.02 (s, 1H), 7.79–7.70 (m, 1H), 6.83 (br, s, 2H), 6.25–6.18 (m, 1H), 5.35 (d, J = 4.31 Hz, 1H), 4.99 (br, s, 1H), 4.38 (br, s, 1H), 3.92 (d, J = 6.42 Hz, 1H), 3.78 (br, s, 1H), 3.63–3.55 (m, 1H), 3.50–3.38 (m, 1H), 3.30–3.20 (m, 4H), 3.05–2.97 (m, 1H), 2.68–2.58 (m, 2H), 2.56–2.49 (m, 4H), 2.12–0.98 (m, 32 H), 0.91 (s, 3H), 0.84 (d, J = 6.41 Hz, 3H), 0.59 (s, 3H); 13C-NMR: δ = 172.6 (q), 156.5 (q), 154.3 (q), 151.0 (q), 146.7 (q), 132.2 (q), 130.2 (q), 120.3 (CH), 117.2 (q), 95.7 (q), 88.1 (CH), 84.1 (CH), 71.6 (CH), 69.6 (q), 69.5 (CH), 62.5 (CH2), 60.1 (CH), 55.7 (CH), 55.1 (CH), 51.1 (CH2), 43.5 (CH), 43.03 (CH), 38.8 (CH), 37.7 (CH2), 37.4 (CH2), 35.9 (CH2), 35.5 (CH2), 35.3 (CH), 34.6 (CH2), 34.2 (q), 33.0 (CH2), 32.9 (CH2), 32.0 (CH2), 28.6 (CH2), 28.1 (CH2), 27.6 (CH2), 27.1 (CH2), 25.1 (CH2), 23.6 (CH3), 22.5 (CH2), 21.4 (CH2), 18.9 (CH3), 18.8 (CH2), 12.5 (CH3); HRMS calculated for [C44H65N9O9S + H]+ 896.4698, found 896.4703.
G-TUDCA. Yield 60%; IR: ν (cm−1) 3329 (O-H), 2933–2867 (C-H), 2254 (C≡C), 1694, 1648 (C=O), 1640–1518 (C=C,C=N); 1H-NMR: δ = 11.13 (s, 1H), 8.02 (s, 1H), 7.75 (br, s, 1H), 6.85 (s, 2H), 5.75 (d, J = 6.10 Hz, 1H), 5.43 (d, J = 6.11 Hz, 1H), 5,11 (d, J = 4.89 Hz, 1H), 4.92–4.97 (m, 1H), 4.88 (q, J = 5.80 Hz, 1H), 4.38 (br, s, 1H), 4.11 (br, s, 1H), 3.92 (d, J = 6.41 Hz, 1H), 3.82 (br, s, 1H), 3.65–3.58 (m, 1H), 3.52–3.38(m, 1H), 3.33–3.23 (m, 4H), 2.66–2.63 (m, 2H), 2.56–2.49 (m, 4H), 2.09–0.98 (m, 30H), 0.92 (s, 3H), 0.86 (d, J = 6.41 Hz, 3H), 0.59 (s, 3H); 13C-NMR: δ = 172.2 (q), 155.9 (q), 154.1 (q), 150.7 (q), 146.2 (q), 130.1 (q), 129.5 (q) 119.9 (CH), 116.9 (q), 95.1 (q), 88.4 (CH), 85.5 (CH), 71.1 (q), 70.8 (CH), 70.6 (CH), 69.0 (CH), 62.0 (CH2), 59.6 (CH), 55.3 (CH), 54.6 (CH), 50.6 (CH2), 43.0 (CH), 42.6 (q), 38.3 (CH2), 38.2 (CH), 37.3 (CH2), 37.2 (CH), 35.42 (CH2), 34.9 (CH2), 34.1 (q), 33.8 (CH2), 32.6 (CH2) 32.5 (CH2), 31.5 (CH2), 28.2 (CH2), 27.7 (CH2), 27.2 (CH2), 26.7 (CH2), 24.6 (CH2), 23.17 (CH3), 20.9 (CH2), 18.5 (CH3), 18.4 (CH2), 12.0 (CH3); HRMS calculated for [C44H65N9O10S + H]+ 912.4647, found 912.4650.
dU-TUDCA. Yield 65%; IR: ν (cm−1) 3442 (O-H), 3015–2860 (C-H), 1690, 1668 (C=O), 1640–1520 (C=C,C=N); 1H-NMR: δ = 8.12 (s, 1H), 8.01 (s, 1H), 7.69–7.61 (m, 2H), 6.32–6.18 (m, 1H), 5.23 (d, J = 4.32 Hz, 1H), 5.19–5.11 (m, 1H), 4.42–4.32 (m, 1H), 4.28–4.19 (m, 2H), 3.91 (d, J = 6.40 Hz, 1H), 3.78 (br, s, 1H), 3.63–3.55 (m, 2H), 3.50–3.38 (m, 2H), 3.30–3.20 (m, 4H), 3.11–3.02 (m, 2H), 2.65–2.54 (m, 2H), 2.42–2.34 (m, 2H), 2.12–1.08 (m, 29 H), 0.92 (s, 3H), 0.88 (d, J = 6.41 Hz, 3H), 0.61 (s, 3H); 13C-NMR: δ = 175.4 (q), 172.60 (q), 161.9 (q), 149.3 (q), 146.6 (q), 143.2 (CH), 120.3 (CH), 99.4 (q), 93.5 (q), 88.0 (CH), 85.0 (CH), 80.7 (CH), 71.6 (CH), 69.6 (CH), 61.4 (CH2), 60.1 (CH), 55.1 (CH), 51.1 (CH), 43.5 (CH), 43.0 (q), 42.6 (CH2), 40.0 (CH2), 38.8 (CH), 37.74 (CH2), 35.5 (CH2), 35.3 (CH), 34.6 (CH2), 33.6 (q), 33.8 (CH2), 33.0 (CH2), 32.9 (CH2), 28.6 (CH2), 28.1 (CH2), 27.7 (CH2), 27.1 (CH2), 26.7 (CH2), 25.1 (CH3), 24.6 (CH2), 23.6 (CH2), 21.4 (CH2), 18.9 (CH3), 18.8 (CH2), 12.4 (CH3); MS (ESI, ES+) m/z: 918 (M + 23).