Functional Characterization of a Hydroxyacid/Alcohol Hydroxycinnamoyl Transferase Produced by the Liverwort Marchantia emarginata
Abstract
:1. Introduction
2. Results and Discussion
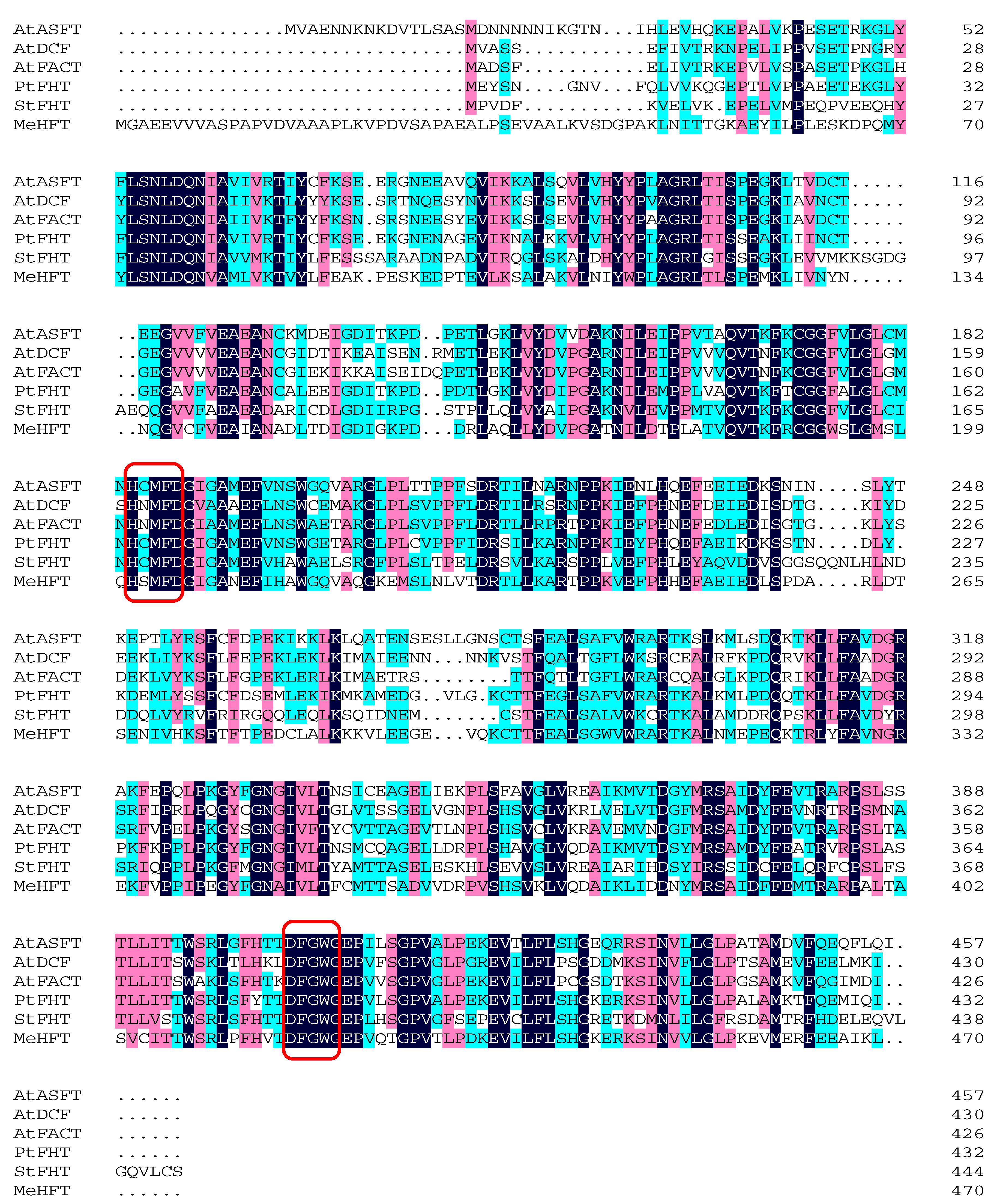
2.1. Gene Isolation and Sequence Analysis
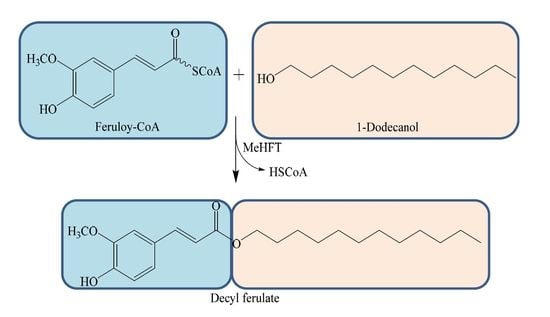
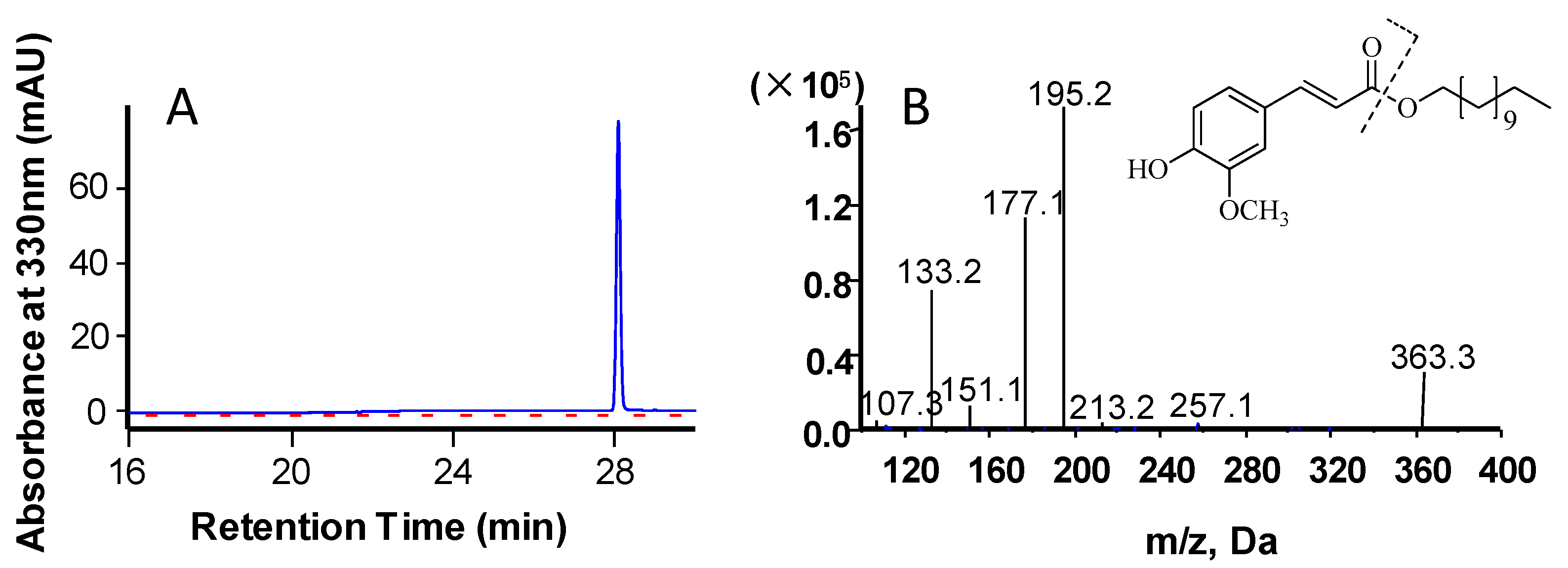
2.2. Functional Analysis
2.3. The Subcellular Localization of MeHFT
3. Materials and Methods
3.1. Plant Materials
3.2. Reagents
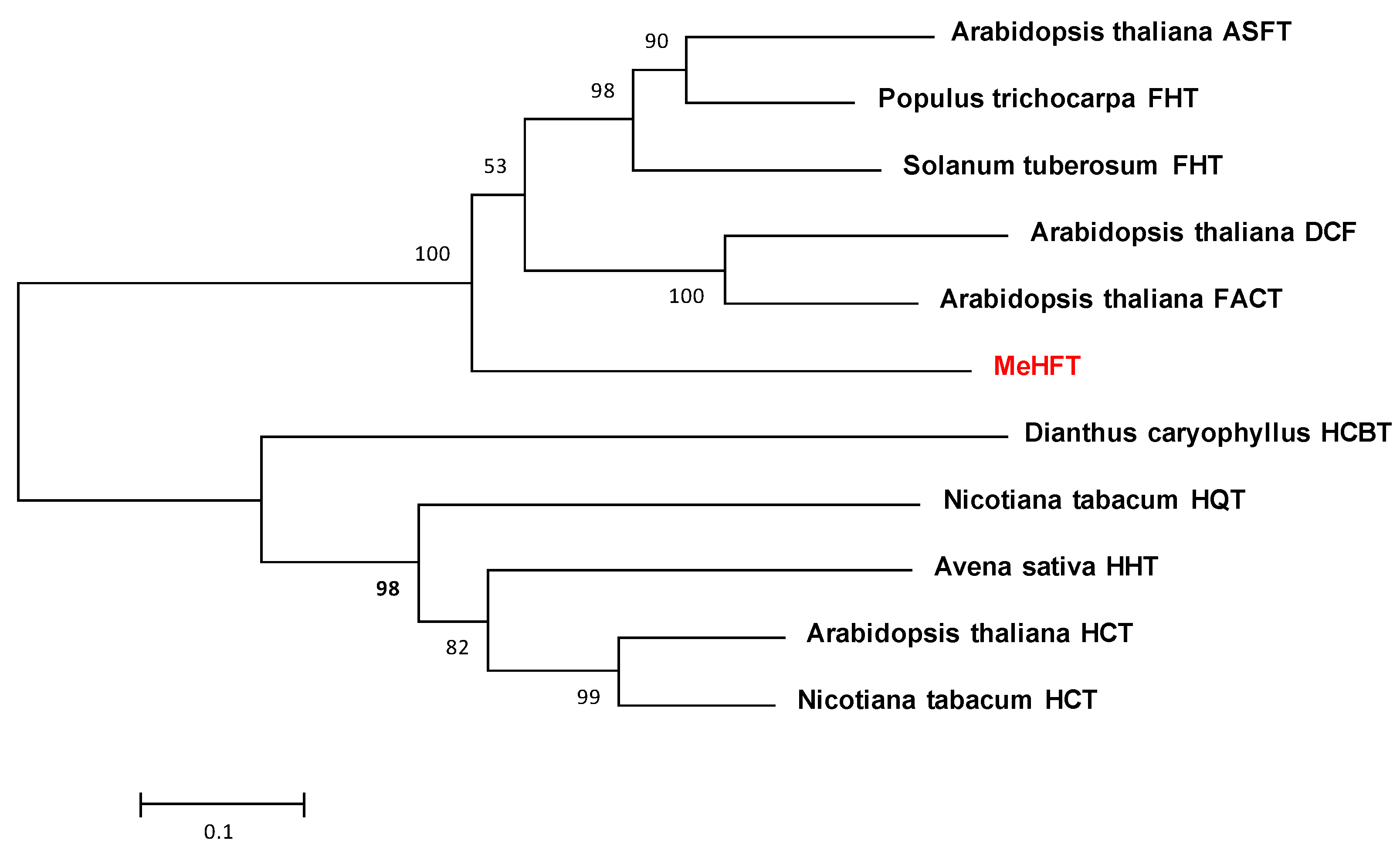
3.3. Phylogenetic Analysis of the MeHFT Sequence
3.4. Gene Isolation, Recombinant Protein Expression and Purification
3.5. Enzymatic Assay and Kinetics Determination
3.6. The Subcellular Localization of MeHFT
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pollard, M.; Beisson, F.; Li, Y.; Ohlrogge, J.B. Building lipid barriers: Biosynthesis of cutin and suberin. Trends Plant Sci. 2008, 13, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, L. Transport barriers made of cutin, suberin and associated waxes. Trends Plant Sci. 2010, 15, 546–553. [Google Scholar] [CrossRef] [PubMed]
- Beisson, F.; Li-Beisson, Y.; Pollard, M. Solving the puzzles of cutin and suberin polymer biosynthesis. Curr. Opin. Plant Biol. 2012, 15, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Franke, R.; Schreiber, L. Suberin—A biopolyester forming apoplastic plant interfaces. Curr. Opin. Plant Biol. 2007, 10, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Graca, J.; Santos, S. Suberin: A biopolyester of plants’ skin. Macromol. Biosci. 2007, 7, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Panikashvili, D.; Shi, J.X.; Schreiber, L.; Aharoni, A. The Arabidopsis DCR encoding a soluble BAHD acyltransferase is required for cutin polyester formation and seed hydration properties. Plant Physiol. 2009, 151, 1773–1789. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M. Hydroxycinnamoyltransferases in plant metabolism. Phytochem. Rev. 2015, 15, 699–727. [Google Scholar] [CrossRef]
- Ma, X.; Koepke, J.; Panjikar, S.; Fritzsch, G.; Stockigt, J. Crystal structure of vinorine synthase, the first representative of the BAHD superfamily. J. Biol. Chem. 2005, 280, 13576–13583. [Google Scholar] [CrossRef] [PubMed]
- St-Pierre, B.; De Luca, V. Evolution of Acyltransferase Genes: Origin and Diversification of the BAHD Superfamily of Acyltransferases Involved in Secondary Metabolism. Recent Adv. Phytochem. 2000, 34, 285–315. [Google Scholar]
- D’Auria, J.C. Acyltransferases in plants: A good time to be BAHD. Curr. Opin. Plant Biol. 2006, 9, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Dudareva, N.; Pichersky, E. Biochemical and molecular genetic aspects of floral scents. Plant Physiol. 2000, 122, 627–633. [Google Scholar] [CrossRef] [PubMed]
- D’Auria, J.C.; Chen, F.; Pichersky, E. Characterization of an acyltransferase capable of synthesizing benzylbenzoate and other volatile esters in flowers and damaged leaves of Clarkia breweri. Plant Physiol. 2002, 130, 466–476. [Google Scholar] [CrossRef] [PubMed]
- D’Auria, J.C.; Pichersky, E.; Schaub, A.; Hansel, A.; Gershenzon, J. Characterization of a BAHD acyltransferase responsible for producing the green leaf volatile (Z)-3-hexen-1-yl acetate in Arabidopsis thaliana. Plant J. 2007, 49, 194–207. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Nakayama, T.; Yonekura-Sakakibara, K.; Fukui, Y.; Nakamura, N.; Nakao, M.; Tanaka, Y.; Yamaguchi, M.A.; Kusumi, T.; Nishino, T. Malonyl-CoA:anthocyanin 5-O-glucoside-6‴-O-malonyltransferase from scarlet sage (Salvia splendens) flowers. Enzyme purification, gene cloning, expression, and characterization. J. Biol. Chem. 2001, 276, 49013–49019. [Google Scholar] [CrossRef] [PubMed]
- D’Auria, J.C.; Reichelt, M.; Luck, K.; Svatos, A.; Gershenzon, J. Identification and characterization of the BAHD acyltransferase malonyl CoA: Anthocyanidin 5-O-glucoside-6′′-O-malonyltransferase (At5MAT) in Arabidopsis thaliana. FEBS Lett. 2007, 581, 872–878. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Nishiyama, Y.; Fuell, C.; Taguchi, G.; Elliott, K.; Hill, L.; Tanaka, Y.; Kitayama, M.; Yamazaki, M.; Bailey, P.; et al. Convergent evolution in the BAHD family of acyl transferases: Identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana. Plant J. 2007, 50, 678–695. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.H.; Chen, M.H.; Liu, C.J. Nucleocytoplasmic-localized acyltransferases catalyze the malonylation of 7-O-glycosidic (iso)flavones in Medicago truncatula. Plant J. 2008, 55, 382–396. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, G.; Ubukata, T.; Nozue, H.; Kobayashi, Y.; Takahi, M.; Yamamoto, H.; Hayashida, N. Malonylation is a key reaction in the metabolism of xenobiotic phenolic glucosides in Arabidopsis and tobacco. Plant J. 2010, 63, 1031–1041. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Fuell, C.; Parr, A.; Hill, L.; Bailey, P.; Elliott, K.; Fairhurst, S.A.; Martin, C.; Michael, A.J. A novel polyamine acyltransferase responsible for the accumulation of spermidine conjugates in Arabidopsis seed. Plant Cell 2009, 21, 318–333. [Google Scholar] [CrossRef] [PubMed]
- Gou, J.Y.; Yu, X.H.; Liu, C.J. A hydroxycinnamoyltransferase responsible for synthesizing suberin aromatics in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 18855–18860. [Google Scholar] [CrossRef] [PubMed]
- Molina, I.; Li-Beisson, Y.; Beisson, F.; Ohlrogge, J.B.; Pollard, M. Identification of an Arabidopsis feruloyl-coenzyme A transferase required for suberin synthesis. Plant Physiol. 2009, 151, 1317–1328. [Google Scholar] [CrossRef] [PubMed]
- Lotfy, S.; Javelle, F.; Negrel, J. Distribution of hydroxycinnamoyl-CoA: ω-hydroxypalmitic acid O -hydroxycinnamoyltransferase in higher plants. Phytochemistry 1995, 40, 389–391. [Google Scholar] [CrossRef]
- Lotfy, S.; Javelle, F.; Negrel, J. Purification and characterization of hydroxycinnamoyl-Coenzyme A: ω-hydroxypalmitic acid O-hydroxycinnamoyltransferase from tobacco (Nicotiana tabacum L.) cell-suspension cultures. Planta 1996, 199, 475–480. [Google Scholar]
- Serra, O.; Hohn, C.; Franke, R.; Prat, S.; Molinas, M.; Figueras, M. A feruloyl transferase involved in the biosynthesis of suberin and suberin-associated wax is required for maturation and sealing properties of potato periderm. Plant J. 2010, 62, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Rautengarten, C.; Ebert, B.; Ouellet, M.; Nafisi, M.; Baidoo, E.E.; Benke, P.; Stranne, M.; Mukhopadhyay, A.; Keasling, J.D.; Sakuragi, Y.; et al. Arabidopsis Deficient in Cutin Ferulate encodes a transferase required for feruloylation of omega-hydroxy fatty acids in cutin polyester. Plant Physiol. 2012, 158, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.X.; Gou, J.Y.; Yu, X.H.; Yang, H.; Fang, X.; Chen, X.Y.; Liu, C.J. Characterization and ectopic expression of a populus hydroxyacid hydroxycinnamoyltransferase. Mol. Plant 2013, 6, 1889–1903. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, 585–587. [Google Scholar] [CrossRef] [PubMed]
- Rensing, S.A.; Lang, D.; Zimmer, A.D.; Terry, A.; Salamov, A.; Shapiro, H.; Nishiyama, T.; Perroud, P.F.; Lindquist, E.A.; Kamisugi, Y. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 2008, 319, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Asakawa, Y. Terpenoids and Aromatic Compounds with Pharmacological Activity from Bryophytes; Clarendon Press: Oxford, UK, 1990; pp. 369–410. [Google Scholar]
- Espineira, J.M.; Novo Uzal, E.; Gomez Ros, L.V.; Carrion, J.S.; Merino, F.; Ros Barcelo, A.; Pomar, F. Distribution of lignin monomers and the evolution of lignification among lower plants. Plant Biol. 2011, 13, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.X.; Li, G.Y.; Wang, F.Q. A cytotoxic diterpenoid and antifungal phenolic compounds from Frullania muscicola Steph. J. Asian Nat. Prod. Res. 2002, 4, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Asakawa, Y. Recent advances in phytochemistry of bryophytes-acetogenins, terpenoids and bis(bibenzyl)s from selected Japanese, Taiwanese, New Zealand, Argentinean and European liverworts. Phytochemistry 2001, 32, 297. [Google Scholar] [CrossRef]
- Asakawa, Y.; Ludwiczuk, A. Distribution of Cyclic and Acyclic Bis-bibenzyls in the Marchantiophyta (Liverworts), Ferns and Higher Plants and Their Biological Activities, Biosynthesis, and Total Synthesis. Heterocycles 2012, 86, 891–917. [Google Scholar] [CrossRef]
- Kosma, D.K.; Molina, I.; Ohlrogge, J.B.; Pollard, M. Identification of an Arabidopsis fatty alcohol: caffeoyl-Coenzyme A acyltransferase required for the synthesis of alkyl hydroxycinnamates in root waxes. Plant Physiol. 2012, 160, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Yu, H.N.; Xu, R.X.; Cheng, A.X.; Lou, H.X. Cloning and functional characterization of a 4-coumarate CoA ligase from liverwort Plagiochasma appendiculatum. Phytochemistry 2015, 111, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Beuerle, T.; Pichersky, E. Enzymatic synthesis and purification of aromatic coenzyme a esters. Anal. Biochem. 2002, 302, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Gambino, G.; Perrone, I.; Gribaudo, I. A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 2008, 19, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wu, Y.F.; Zhao, Y.; Han, X.J.; Lou, H.X.; Cheng, A.X. Molecular cloning and biochemical characterization of two cinnamyl alcohol dehydrogenases from a liverwort Plagiochasma appendiculatum. Plant Physiol. Biochem. 2013, 70, 133–141. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |


| Chemicals | Specific Activity (%) | Km (μM) | Vmax (nmol mg−1 min−1) | Kcat (s−1) | Kcat/Km (M−1s−1) |
|---|---|---|---|---|---|
| Acyl donor | |||||
| Feruloyl-CoA | 100* | 79.4 ± 10.86 a 83.23 ± 18.70 b | 83.23 ± 18.70 a 290 ± 37.49 b | 0.25263945 ± 0.015 a 0.250995 ± 0.032 b | 3181 a 3015 b |
| p-Coumaroyl-CoA | 38 | ||||
| Caffeoyl-CoA | 43 | ||||
| Acyl acceptor | |||||
| 1-Hexanol | 31 | ||||
| 1-Heptanol | 50 | ||||
| 1-Decanol | 100 * | 65.5 ± 11.21 | 297.3 ± 21.04 | 0.25731315 ± 0.018 | 3928 |
| 1-Dodecanol | 89 | ||||
| 1-Tetradecanol | 85 | ||||
| 1-Hexadecanol | n.d. | ||||
| 16-Hydroxypalmitic acid | 71 | ||||
| Palmitic acid | n.d. | ||||
| 1-Eicosanol | n.d. | ||||
| 1-Docosanol | n.d. |
| Primer Name | Primer Sequences (5’ to 3’) |
|---|---|
| MeHFT-F | ATGGGCGCCGAGGAAGTG |
| MeHFT-R | CAGCAGGCAGACGGAATT |
| MeHFT-pet32a-F | CGAGCTCATGGGCGCCGAGGAAGTGGT |
| MeHFT-pet32a-R | CCCTCGAGTTACAGCTTGATAGCTTCTT |
| attB1-MeHFT-F | GGGGACAAGTTTGTACAAAAAAGCAGGCTTAACCATGGGCGCCGAGGAAGTGGT |
| attB2-MeHFT-R | GGGGACCACTTTGTACAAGAAAGCTGGGTCCAGCTTGATAGCTTCTTCGA |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.-P.; Liu, H.; Gao, S.; Cheng, A.-X. Functional Characterization of a Hydroxyacid/Alcohol Hydroxycinnamoyl Transferase Produced by the Liverwort Marchantia emarginata. Molecules 2017, 22, 1854. https://doi.org/10.3390/molecules22111854
Wang P-P, Liu H, Gao S, Cheng A-X. Functional Characterization of a Hydroxyacid/Alcohol Hydroxycinnamoyl Transferase Produced by the Liverwort Marchantia emarginata. Molecules. 2017; 22(11):1854. https://doi.org/10.3390/molecules22111854
Chicago/Turabian StyleWang, Ping-Ping, Hui Liu, Shuai Gao, and Ai-Xia Cheng. 2017. "Functional Characterization of a Hydroxyacid/Alcohol Hydroxycinnamoyl Transferase Produced by the Liverwort Marchantia emarginata" Molecules 22, no. 11: 1854. https://doi.org/10.3390/molecules22111854