Abstract
Lichens are symbiotic associations of fungi with microalgae and/or cyanobacteria. Lichens belonging to the Parmeliaceae family comprise 2700 species of lichens, including the Parmotrema genus which is composed of 300 species. The metabolites of this genus include depsides, depsidones, phenolics, polysaccharides, lipids, diphenylethers and dibenzofurans, which are responsible for the biological activities reported including antidiabetic, antihelmintic, anticancer, antioxidant, antibacterial, anti-inflammatory, antimitotic, antitumoral, antifungal, and antioxidant enzyme inhibitory. Due to scarce knowledge of metabolomic profiles of Parmotrema species (P. andinum and P. robustum), a full metabolome study based on ultra-high performance liquid chromatography- diode array detector-electrospray ionization-quadrupole-orbitrap-mass-spectrometry (UHPLC-DAD-ESI-Q-orbitrap MS) was performed for a comprehensive characterization of their substances. From the methanolic extracts of these species, a total of 54 metabolites were identified for the first time using this hyphenated technique, including thirty compounds in P. andinum, and thirty-seven in P. robustum. Moreover, two compounds were not identified as known compounds, and could be new structures, according to our data. This report shows that this technique is effective and accurate for rapid chemical identification of lichen substances and the compounds identified could serve as chemotaxonomic markers to differentiate these ruffle lichens.
1. Introduction
Lichens, by definition, correspond to a symbiotic association between a fungus and one or more photosynthetic autotrophic organisms that may be a green algae or a cyanobacterium, resulting in a morphologically different thallus to each of its components as a totally new morphological entity [1] Recently it was discovered that a third party, such as basidiomicete yeast, can also be a component of lichens [2]. In the Colombian Andean region, 1396 species of lichens have been found distributed in 295 genera and 88 families, representing 89% of the species, 95% of the genera and 93% of the families. The richest families are Parmeliaceae, Graphidaceae, Physciaceae, Thelotremataceae and Cladoniaceae and the most diversified genera are represented by Cladonia, Parmotrema, Hypotrachyna, Usnea, Arthonia, Porina, Lecanora, Leptogium and Sticta [3]. Some 215 genera of lichens have been reported in Peru; The genus with the highest number of recorded species is Cladonia, followed by Hypotrachyna and Heterodermia, and the species with the greatest presence are Chrysothrix candelaris and Cladonia melanopoda [3].
The genus Parmotrema (spp.) is characterized by having flat lobes of the thallus growing along the substrate, often largely adhered to it. The thallus shows similar structures to threads, more or less branched, in the ventral face (rhizinas) or in the margin (cilia). Rhizines are not dichotomically branched. The lobes of the thallus have a size of between 0.2 cm and more than 1 cm wide, with colorless, simple or bacillary spores and a ventral face devoid of veins [1]. Parmotrema lichens are difficult to differentiate since they are similar in appearance [4,5,6,7,8], thus, the chemotaxonomical differentiation of these lichens is very important. Some chemical studies of Parmotrema species have been reported previously and some compounds were identified [5,9,10,11,12,13,14,15,16,17,18]. The compound 2-methylene-3-(R)-hydroxynonadecanoic acid was reported from P. xanthinum [5,12,13], while praesorediosic acid and protopraesorediosic acid were reported from P. praesorediosum [16,17], and methyl β-orcinolcarboxylate, atranorin, isolecanoric acid and lecanoric acid from P. tinctorum [18]. Furthermore, the depside 4-O-demethylmicrophyllinic acid was reported from P. demethylmicrophyllinicum [9,10], and protocetraric acid was reported from P. dilatatum [5,12,13]. Then consalazinic acid was found in P. subisidiosum, malonprotocetraric acid in P. conformatum [14], the depside β-alectoronic acid was reported from Parmotrema sp., and finally salazinic acid, methyl orsellinate and orsellinic acid were reported to occur in P. stuppeum [15].
Regarding the biological activity of Parmotrema species, several organic extracts of P. grayana, P. pseudotinctorum, P. praesorediosum, P. stuppeum, P. tinctorum, and Parmotrema sp. have been assayed for antidiabetic, antihelmintic, anticancer, antioxidant, antibacterial, anti-inflammatory, antimitotic, antitumoral, antifungal, and enzyme inhibitory activity [11].
High performance liquid chromatography-tandem mass spectrometry (HPLC-MS-MS) is a powerful technique that combines liquid chromatography with mass analyses and whose application is the detection and identification of metabolites in complex extracts, including unknown metabolites, based on its fragmentation patterns. A pioneering work by Leuckert and Holzmann [19] detected lichen substances using fast atom bombardment MS-MS and identified usnic acid, diffractaic acid, gyrophoric acid, lecanoric acid, orsellinic acid, ovoic acid, thamnolic acid, hypothamnolic acid, divaricatic acid, fumarprotocetraric acid, protocetraric acid, homosekikaic acid and sekikaic acid on the following lichens: Alectoria ochroleuca, Umbilicaria torrefacta, Thamnolia vermicularis, Ophioparma ventosa, Cladonia cryptochlorophaea and Cladonia rei [19]. Later, Parrot et al. [20] reported a study of eight chemotypes of Ramalina siliquosa using LC-ESI-MS-MS and identified ten lichen substances: conhypoprotocetraric acid, salazinic acid, peristictic acid, cryptostictic acid, protocetraric acid, stictic acid, norstictic acid, hypoprotocetraric acid, 4-O-demethylbarbatic acid, usnic acid. In another report, β-orcinol, orsenillic acid, choline sulphate, roccellic acid, montagnetol, lecanoric acid, erythrin, lepraric acid and acetylportentol were identified based on the HPLC–MS–MS approach in nine lichens belonging to the Lichina, Collema and Roccella genera [21]. Afterwards, Le Pogam et al. [22] proposed the rapid identification of lichen extracts using laser desorption/ionization time of flight mass spectrometry instead of electrospray ionization. The analyzed samples with this MS technique were Diploicia canescens, Evernia prunastri, Ophioparma ventosa, Pseudevernia furfuracea, Roccella fuciformis, Xanthoria parietina, Cladonia portentosa, flavocetraria nivalis, Lecidella asema, Ramalina siliquosa, Vulpicida pinastri and Usnea filipendula. However, only 2 to 5 compounds were reported in each studied species in general [22]. Finally, the lichens Parmotrema grayana and Heterodermia obscurata were studied using high performance liquid chromatography- electrospray ionization-quadrupole-time of flight-mass-mass spectrometry (HPLC-ESI-Qq-TOF-MS-MS) on negative ion mode and fifteen compounds were detected and identified from the organic extracts [23].
The Q-Exactive Focus is a hybrid high-resolution mass spectrometer used to detect and quantify small organic compounds. The hyphenated Q-Exactive Focus instrument is an HRAM instrument (high resolution accurate mass) which combines UHPLC-DAD (UHPLC-diode array detection) with an orbital trap (orbitrap), a quadrupole (Q) and a high-resolution collision cell (HCD), which allows high resolution diagnostic MS fragments [22,23,24,25,26]. In a continuation of our research on the identification of lichen substances [25,26], we have selected two unstudied Parmotrema lichens for chemotaxonomic fingerprinting and describe the full comprehensive phytochemical profile of P. andinum and P. robustum for the first time, based on UHPLC-DAD coupled with high resolution electrospray ionization tandem mass spectrometry (UHPLC-Q-Orbitrap-HRMS). The profiles serve as fingerprints to differentiate these lichens since the ruffle lichens are difficult to differentiate.
2. Results and Discussion
Electrospray orbitrap emerged as a very fast and versatile tool for the rapid identification of lichens [25,26]. Thus, two Parmotrema species were selected, P. andinum from Ancash, Peru and P. robustum from Colombia, in order to determine their metabolomic profiles and chemical fingerprints in order to differentiate them since the ruffle lichens are similar in appearance and very difficult to differentiate [4,6,8,27]. Below is the detailed explanation of the rapid metabolome analysis of the aforementioned unstudied Parmotrema species using this HRAM technique.
2.1. Parmotrema andinum
Thirty compounds (Figure 1) were detected for the first time in a methanolic extract using UHPLC-ESI-MS-MS in negative mode (Table 1, Table S1). The identified compounds were mainly depsides, depsidones, lipids, aromatics, diphenylethers and dibenzofurans [9,10,22,23,24,25,26].
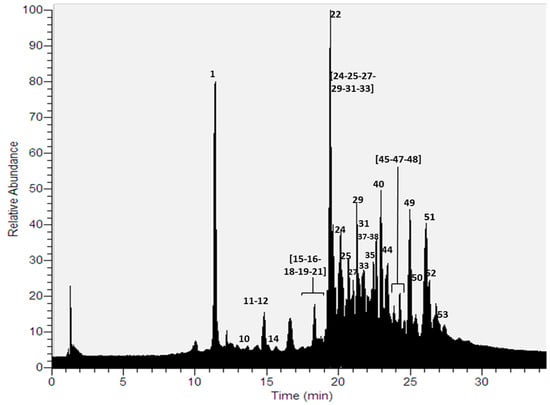
Figure 1.
UHPLC Chromatogram of P. andinum.

Table 1.
Identification of lichen substances in Parmotrema species by UHPLC-ESI-MS-MS.
Depsides: Six depsides (peaks 18, 24, 33, 44, 45 and 51) were identified using a combination of diode array detection and high-resolution tandem mass spectrometry. Peak 18 was identified as lecanoric acid, which showed an [M − H]− ion at m/z 317.0668. Major diagnostic daughter MS ions of lecanoric acid were [M − H − C8H6O3]−, [M − H − C8H8O4]− and [C7H7O2]− (167.0343, 149.0237 and 123.0444 a.m.u., respectively) [25]. Peak 24 was identified as decarboxythamnolic acid (molecular anion at m/z 375.0724), whose fragmentation produced diagnostic MS ions at m/z 209.0450, 167.0345 and 139.0394. Peak 33 was assigned to evernic acid, showing a molecular anion at m/z 331.0825. Its fragmentation produced ions at m/z 167.0345, 149.0238, and 123.0444 a.m.u. Peak 44, with an [M − H]− ion at m/z 429.1922, was identified as 2-O-methylstenosporic acid. The parent ion produced major diagnostic MS ions at m/z 223.0972 [M − H − C11H12O3]−, and 179.1072 [C9H11O2]− confirming this depside. Peak 45 presented a pseudomolecular ion at m/z 359.1138, which produced fragmented ions at m/z 181.0501, 163.0394 and 137.0600, and thus, was identified as barbatic acid. Peak 51 was identified as atranorin, which showed an [M − H]− ion at m/z 373.0930. The major diagnostic daughter ions were at m/z 177.0190 and 163.0386 a.m.u.
Depsidones: Seven depsidones corresponding to peaks 11, 15, 16, 22, 25, 35, and 48 were identified using UHPLC-DAD and HRMS-MS analysis [25]. Peak 11 was identified as salazinic acid, which showed an [M − H]− ion at m/z 387.0359. Its major diagnostic daughter ions were at m/z 243.0378, 227.0343, 151.0394 and 121.0291 a.m.u. Peaks 15 and 16 were identified as stictic acid and connorstictic acid, which showed [M − H]− ions at m/z 385.0568 and 373.0568 respectively. The major diagnostic daughter ions were at m/z 341.0668, 297.0760, 267.0297 and 165.0544 a.m.u. for stictic acid, while for connorstictic acid ions they were at m/z 329.0665, and 181.0554 a.m.u. Peak 22, with an [M − H]− pseudomolecular ion at m/z 371.0411, was identified as substictic acid, which showed diagnostic daughter ions at m/z 327.0512, and 255.0660. Hypoconstictic acid was identified as peak 25 (molecular anion at m/z 387.0724). The fragmentation of peak 25 produced ions at m/z 343.0825, and 299.0921. Peak 35, with an [M − H]− ion at m/z 551.1194, was identified as furfuric acid. The parent ion produced major diagnostic MS ions at m/z 359.1130, 179.0345, 163.0394 and 137.0601 confirming this compound. Finally, peak 48 was identified as lobaric acid (molecular anion at m/z 455.1712) [26]. The fragmentation of peak 48 also produced ions at m/z 411.1815 [M − H − CO2]−, 367.1909 [M − H − 2CO2]−, 352.1681 [M − H − 2CO2 − CH3]−, and 296.1048 [M − H − 2CO2 − C5H11]− confirming this depsidone.
Lipids: Six polyhydroxylated lipids were tentatively identified (peaks 21, 29, 38, 40, 49 and 53) using UHPLC–ESI–MS–MS analysis. Peak 21, with an [M − H]− ion at m/z 403.3069, was tentatively identified as tetrahydroxydocosanoic acid. Peak 29 showed an [M − H]− ion at m/z 517.3748 and was tentatively identified as pentahydroxyoxooctacosanoic acid. Peaks 38, 40 and 49 were tentatively identified as hydroxydioxohenicosanoic acid (C21H38O5), trioxohenicosanoic acid (C21H36O5) and dihydroxydioxononadecanoic (C19H34O6), which showed [M − H]− ions at m/z 369.2649, 367.2492 and 357.2285 respectively. Finally, peak 53, with an [M − H]− ion at m/z 295.1917, was tentatively identified as dihydroxyheptadecatrienoic acid (C17H28O4) [28].
Diphenylethers: Three diphenylethers (peak 27, 47 and 52) were detected in the methanolic extract using UHPLC-DAD-MS-MS analysis. Peak 27 was identified as loxodinol, which showed an [M − H]− ion at m/z 473.1820. Its major diagnostic daughter ions were at m/z 237.1126, and 221.0819 a.m.u. Peak 47 and peak 52 were identified as α-collatolic acid and β-collatolic acid [29], which showed [M − H]− ions at m/z 525.2130 for both. Their major diagnostic daughter ions were at m/z 263.1281 and 265.1076 a.m.u respectively.
Dibenzofurans: Strepsilin, with an [M − H]− ion at m/z 269.0455, was evidenced as peak 12 [30]. The main daughter ions of peak 12 were at m/z 149.0238 and 123.0450 a.m.u. Peak 50 was identified as usnic acid [25] (molecular ion at m/z 343.0824). The main daughter ions of this peak were [M − H − CH3]−, [M − H − C4H3O2]− and [M − H − C5H3O3]− (328.0591, 259.0609 and 231.0661 a.m.u., respectively).
Aromatic compounds: Five simple aromatic compounds corresponding to the peaks 1, 10, 14, 19 and 37 were identified using UHPLC-DAD and HRMS–MS analysis. Peak 1 was identified as orsellinic acid [25], which showed an [M − H]− ion at m/z 167.0345. The major diagnostic daughter MS ion of this compound was at m/z 123.0445 a.m.u. Peak 10 was identified as atranol (molecular anion at m/z 151.0395) whose fragmentation produced a diagnostic MS ion at m/z 123.0444. Peak 14 was assigned to haematommic acid [31] whose molecular anion was at m/z 195.0296. Its fragmentation produced ions at m/z 151.0390, 149.0240, and 123.0440 a.m.u. Peak 19 was identified as pentyldivaric acid, which showed an [M − H]− ion at m/z 223.0974. Major diagnostic daughters MS ions of this peak were at m/z 167.0344, 149.0238 and 123.0445 a.m.u. Finally, ethylhaematommate was assigned to peak 37 (molecular anion at m/z 223.0610) whose fragmentation produced diagnostic MS ions at m/z 177.0189, 149.0238 and 123.0444 a.m.u.
Chromones: Lepraric acid (peak 31) was detected in the methanolic extract using this hyphenated technique [32]. Peak 31 showed an [M − H]− ion at m/z 361.0931. Its major diagnostic daughter ions were at m/z 235.0606, 195.0292 and 149.0236 a.m.u.
2.2. Parmotrema Robustum
Thirty-seven compounds (Figure 2) were detected for the first time in a methanolic extract of this species using UHPLC-ESI-MS-MS in negative mode (Table 1, Supplementary Materials Table S1). As in the previous case, the compounds were mainly depsides, depsidones, lipids, aromatics, diphenylethers and dibenzofurans [9,10,22,23,24,25,26].
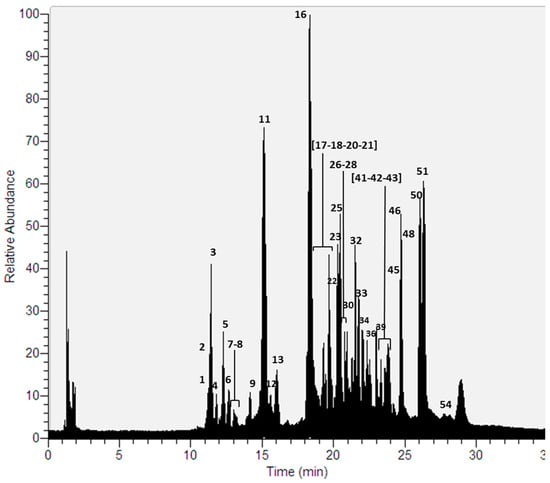
Figure 2.
UHPLC Chromatogram of P. robustum.
Depsides: Thirteen depsides (peaks 6, 7, 9, 18, 30, 33, 39, 42, 43, 45, 46, 51 and 54) were identified. Peak 6 and 7 were identified as thamnolic acid and haemathamnolic acid, which showed [M − H]− ions at m/z 419.0623 and 403.0675 respectively. Thamnolic acid produced major diagnostic MS ions at m/z 375.0722 and 167.0345 while haemathamnolic acid produced ions at m/z 209.0002 and 193.0503 u.m.a. Peak 30, with an [M − H]− pseudomolecular ion at m/z 467.0984, was identified as gyrophoric acid, which showed diagnostic daughter ions at m/z 317.0667, 167.0344, 149.0240 and 123.0444. Peak 42 was identified as 4-O-methylgyrophoric acid based on its pseudomolecular ion at m/z 481.1141 and its daughter ions at m/z 317.0668, 167.0343, 149.0240 and 123.0443. Pseudocyphellarin A was assigned to peak 43 (molecular anion at m/z 401.1244). Major diagnostic daughter MS ions were at m/z 191.0347, 177.0552 and 133.0651 a.m.u. Peak 46 was identified as sekikaic acid (molecular anion at m/z 417.1559). The fragmentation of peak 46 produced ions at m/z 225.0768 [M − H − C11H12O3]−, 209.0814 [M − H − C11H12O4]−, and 165.0915 [M − H − C12H12O5]−. Peak 54 was identified as chloroatranorin [33], which showed an [M − H]− ion at m/z 407.0542. The major diagnostic daughter ions were at m/z 228.9906, 210.9800 and 163.0394 a.m.u. Finally, peaks 18, 33, 45, and 51 were identified as lecanoric acid, evernic acid, barbatic acid and atranorin respectively.
Depsidones: Thirteen depsidones corresponding to the peaks 2, 4, 5, 8, 11, 13, 16, 17, 22, 23, 25, 41 and 48 were identified in this species. Peak 2 was identified as consalazinic acid [34] which showed an [M − H]− ion at m/z 389.0517. Its major diagnostic daughter ions were at m/z 371.0479, 309.0406, 253.0506 and 209.0605 a.m.u. Peaks 4, 8 and 13 were identified as conzalasinic acid derivatives based on both their pseudomolecular ions and daughter ions. Conprotocetraric acid [35] was at peak 5 (molecular anion at m/z 375.0724) and their fragmentation produced ions at m/z 357.0610, 313.0722, 295.0618 and 251.0710. Peak 17, with a [M − H]− ion at m/z 401.0516, was identified as a constictic acid derivative. The parent ion produced major diagnostic MS ions at m/z 373.0568, 357.0618, and 151.0392, confirming this derivative, unlike constictic acid whose daughter ions were at m/z 373.0565, 357.0614, 283.0601, and 227.0698. Peak 23, with a [M − H]− pseudomolecular ion at m/z 371.0412, was identified as norstictic acid [36], which showed diagnostic daughter ions at m/z 283.0616, 267.0667, 243.0292 and 227.0348. α-alectoronic acid was assigned to peak 41 (molecular anion at m/z 511.1973). The fragmentation of peak 41 produced ions at m/z 467.2050, 369.1338 and 247.0974. Finally, peaks 11, 16, 22, 25 and 48 were identified as salazinic acid, connorstictic acid, substictic acid, hypoconstictic and lobaric acid [36], respectively.
Lipids: Five polyhydroxylated lipids were tentatively identified (peaks 20, 21, 26, 28, and 32) using UHPLC–ESI–MS–MS analyses [28]. Peak 20, with an [M − H]− ion at m/z 447.3329, was tentatively identified as pentahydroxytetracosanoic acid. Peaks 26, 28 and 32 showed [M − H]− ions at m/z 431.3380, 475.3635 and 493.3383, and were tentatively identified as tetrahydroxytetracosanoic acid, pentahydroxyhexacosanoic acid and heptahydroxypentacosanoic acid, respectively. As indicated above, peak 21 was identified as tetrahydroxydocosanoic acid.
Diphenylethers: A diphenylether was detected in this species. Peak 36 was identified as β-alectoronic acid [7], which showed an [M − H]− ion at m/z 511.1974. Its major diagnostic daughter ions were at m/z 369.1339, 247.0967 and 163.0396 a.m.u.
Dibenzofurans: Strepsilin (peak 12) and usnic acid (peak 50) were identified in this species, as indicated above.
Aromatic compounds: An aromatic compound corresponding to peak 1 was identified in this analysis. As indicated above, peak 1 was identified as orsellinic acid [26].
Unknown compounds: Two compounds (peaks 3 and 34) were not identified.
Thirteen compounds (Figure 3) were detected in both Parmotrema species, which correspond to orsellinic acid (peak 1), salazinic acid (peak 11), strepsilin (peak 12), connorstictic acid (peak 16), lecanoric acid (peak 18), tetrahydroxydocosanoic acid (peak 21), substictic acid (peak 22), hypoconstictic acid (peak 25), evernic acid (peak 33), barbatic acid (peak 45), lobaric acid (peak 48), usnic acid (peak 50) and atranorin (peak 51). P. robustum produced more depsides and depsidones than P. andinum, while the latter is more a producer of aromatic compounds. Also, in P. andinum a chromone (peak 31) was assigned to lepraric acid. According to SciFinder, there are no chemical studies on P. andinum and P. robustum. Therefore, our work represents the first study on the chemistry of these Parmotrema species.
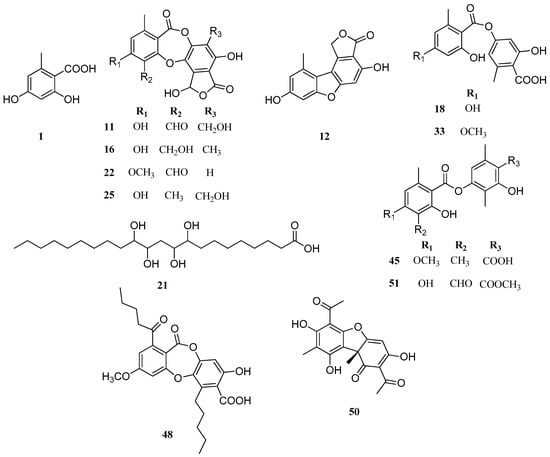
Figure 3.
Chemical structures of similar compounds in P. andinum and P. robustum.
3. Materials and Methods
3.1. Lichen Material
The lichen specimen Parmotrema andinum (Müll. Arg.) Hale (30 g) was collected at “Huaraz,” Ancash, Peru, in 2015. A voucher specimen, PA-61-USM-2015, was deposited in the Museo de Historia Natural de la UNMSM, and Prof. Dr. Haydee Montoya confirmed its identity.
The species Parmotrema robustum (Degel.) (17 g) was collected in “Combeima river basin,” Ibagué-Tolima, Colombia by M. Rivera-Montalvo and Prof. A. Torres-Benítez. A voucher specimen, COL-011, was deposited in the herbarium of Universidad Distrital Francisco José de Caldas, and Prof. Alejandra Suárez Corredor confirmed its identity.
3.2. UHPLC-Orbitrap-ESI-MS-MS
Sample preparation: Some 3 grams of each lichen were macerated with methanol (3 times, 30 mL each time, 3 days/extraction). The solutions were concentrated to obtain 19 mg (P. andinum), and 11 mg (P. robustum) of a gummy extract.
3.2.1. Instrument
A Thermo Scientific Dionex Ultimate 3000 UHPLC system, equipped with a quaternary Series RS pump and Thermo Scientific Dionex Ultimate 3000 Series TCC-3000RS column compartments with a Thermo Fisher Scientific Ultimate 3000 Series WPS-3000RS autosampler and a rapid separations PDA detector controlled by Chromeleon 7.2 Software (Thermo Fisher Scientific, Waltham, MA, USA and Dionex Softron GmbH Part of Thermo Fisher Scientific, Germering, Germany) hyphenated with a Thermo high resolution Q Exactive focus mass spectrometer (Thermo, Bremen, Germany) were used for the analysis. The chromatographic system was coupled to the MS with a Heated Electrospray Ionization Source II (HESI II). Nitrogen (purity > 99.999%) obtained from a Genius NM32LA nitrogen generator (Peak Scientific, Billerica, MA, USA) was employed as both the collision and damping gas. Mass calibration for Orbitrap was performed once a week, in both negative and positive modes, to ensure a working mass accuracy lower than or equal to 5 ppm. Caffeine and N-butylamine (Sigma Aldrich, Saint Louis, MO, USA) were the calibration standards for positive ions and buspirone hydrochloride, sodium dodecyl sulfate, and taurocholic acid sodium salt (Sigma Aldrich, Saint Louis, MO, USA) were used to calibrate the mass spectrometer. These compounds were dissolved in a mixture of acetic acid, acetonitrile, water and methanol (Merck, Darmstadt, Germany) and were infused using a Chemyx Fusion 100 syringe pump (Thermo Fisher Scientific, Bremen, Germany). XCalibur 2.3 software (Thermo Fisher Scientific, Bremen, Germany) and Trace Finder 3.2 (Thermo Fisher Scientific, San José, CA, USA) were used for UHPLC control and data processing, respectively. Q Exactive 2.0 SP 2 from Thermo Fisher Scientific was used to control the mass spectrometer.
3.2.2. LC Parameters
An UHPLC C18 column (Acclaim, 150 mm × 4.6 mm ID, 5 m, Thermo Fisher Scientific, Bremen, Germany) operated at 25 °C was employed. The detection wavelengths were 254, 280, 320 and 440 nm. PDA was recorded from 200 to 800 nm, and mobile phases were 1% formic aqueous solution (A) and acetonitrile (B). The gradient program (time (min), % B) was: (0.00, 5); (5.00, 5); (10.00, 30); (15.00, 30); (20.00, 70); (25.00, 70); (35.00, 5) and 12 min for column equilibration before each injection. The flow rate was 1.00 mL min−1, and the injection volume was 10 µL. Standards and lichen extracts dissolved in methanol were kept at 10 °C inside the autosampler.
3.2.3. MS Parameters
The HESI parameters were as follows: sheath gas flow rate, 75 units; auxiliary gas unit flow rate, 20; capillary temperature, 400 °C; auxiliary gas heater temperature, 500 °C; spray voltage, 2500 V (for ESI-); and S lens, RF level 30. Full scan data in positive and negative were acquired at a resolving power of 70,000 FWHM (full width half maximum) at m/z 200. For the compounds of interest, a scan range of m/z 100–1000 was chosen; the automatic gain control (AGC) was set at 3 × 106 and the injection time was set to 200 ms. The scan-rate was set at 2 scans s−1. External calibration was performed using a calibration solution in positive and negative modes. For confirmation purposes, a targeted MS-MS analysis was performed using the mass inclusion list, with a 30 s time window, with the Orbitrap spectrometer operating both in positive and negative modes at 17,500 FWHM (m/z 200). The AGC target was set to 2 × 105, with the maximum injection time of 20 ms. The precursor ions were filtered by the quadrupole, which operated at an isolation window of m/z 2. The fore vacuum, high vacuum and ultrahigh vacuum were maintained at approximately 2 mbar, from 105 and below 1010 mbar, respectively. Collision energy (HCD cell) was operated at 30 kv. Detection was based on calculated exact mass and on retention time of target compounds, as shown in Table 1. The mass tolerance window was set to 5 ppm for the two modes.
4. Conclusions
In the present report, fifty-four metabolites were detected using UHPLC-Q-Orbitrap-ESI-MS-MS for the first time in P. andinum and P. robustum. Our study indicates that lipids, depsides, depsidones, dibenzofurans, diphenylethers and aromatic compounds were the main compounds detected and identified. This report could contribute to the better understanding of the chemistry of Parmotrema genus, and it supports that the HPLC fingerprints are very important for the fast chemical differentiation of these ruffle lichens.
Supplementary Materials
The following are available online, Table S1: Structure of the compounds identified by UHPLC-ESI-MS-MS from Parmotrema species.
Acknowledgments
This work was supported by Fondecyt Regular N-1170871 and 1150745, INACH RT 16-17, Project 16-443-SEM and 17-477-SEM Universidad de Ibagué-Gobernación del Tolima.
Author Contributions
C.A. and O.G.-B., conceived and designed the experiments; B.S.; O.N.C.; A.T.-B.; M.R.-M.; E.N. and M.J.S. performed the experiments; B.S.; O.G.-B., and M.J.S., analyzed the data; C.A.; O.G.-B., and M.J.S., wrote the paper”.
Conflicts of Interest
The authors declare no conflict of interest. The funding sponsors had no role in the design of the study; in the collection, analyses, interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Chaparro, M.; Aguirre, J. Lichenized Fungi; National University of Colombia-Bogotá: Bogotá, Colombia, 2002. [Google Scholar]
- Spribille, T.; Tuovinen, V.; Resl, P.; Vanderpool, D.; Wolinski, H.; Aime, M.C.; Schneider, K.; Stabentheiner, E.; Toome-Heller, M.; Thor, G.; et al. Basidiomycete yeasts in the cortex of ascomycete macrolichens. Science 2016, 353, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, J. Diversity and richness of the lichens in the Andean natural region or cordilleran system. Wealth and diversity of mosses and lichens in Colombia. In Colombia Biotic Diversity VI; Rangel, O., Ed.; Institute of Natural Sciences: Bogotá, Colombia, 2008; pp. 337–382. [Google Scholar]
- Chen, J.B.; Wang, S.L. Parmeliaceae (Ascomycota) lichens in China’s mainland III. The genus Parmotrema. Mycotaxon 2005, 91, 93–113. [Google Scholar]
- Jayalal, U.; Divakar, P.K.; Joshi, S.; Oh, S.O.; Koh, Y.J.; Hur, J.S. The Lichen Genus Parmotrema in South Korea. Mycobiology 2013, 41, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, S.; Lai, M.J. Parmelioid lichen genera and species in Taiwan. Mycotaxon 2001, 77, 225–284. [Google Scholar]
- Lendemer, J.C.; Allen, J.L.; Noell, N. The Parmotrema acid test: A look at species delineation in the P. perforatum group 40 year later. Mycologia 2015, 107, 1120–1129. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, D.; Bouda, F.; Malicek, J.; Hafellner, J. A contribution to the knowledge of lichenized and lichenicolous fungi in Albania. Herzogia 2012, 25, 149–165. [Google Scholar] [CrossRef]
- Huneck, S.; Yoshimura, I. Identification of Lichen Substances; Springer Verlag: Berlin, Germany, 1996. [Google Scholar]
- Lichen Secondary Metabolites Bioactive properties and Pharmaceutical Potential; Rankovic, B., Ed.; Springer Cham Heidelberg: New York, NY, USA, 2015. [Google Scholar]
- Gomez-Serranillos, M.P.; Fernandez-Moriano, C.; Gonzalez-Burgos, E.; Divakar, P.K.; Crespo, A. Parmeliaceae family: Phytochemistry, pharmacological potential and phylogenetic features. RSC Adv. 2014, 4, 59017–59047. [Google Scholar] [CrossRef]
- Brandao, L.F.G.; Alcantara, G.B.; Matos, M.D.C.; Bogo, D.; Freitas, D.D.; Oyama, N.M.; Honda, N.K. Cytotoxic Evaluation of Phenolic Compounds from Lichens against Melanoma Cells. Chem. Pharm. Bull. 2013, 61, 176–183. [Google Scholar] [PubMed]
- Guterres, Z.D.; Honda, N.K.; Coelho, R.G.; Alcantara, G.B.; Micheletti, A.C. Antigenotoxicity of Depsidones Isolated from Brazilian Lichens. Orbital 2017, 9, 50–54. [Google Scholar] [CrossRef]
- Keogh, M.F. Malonprocetraric acid from Parmotrema conformatum. Phytochemistry 1977, 16, 1102. [Google Scholar] [CrossRef]
- Jayaprakasha, G.K.; Rao, L.J. Phenolic constituents from the lichen Parmotrema stuppeum (Nyl.) Hale and their antioxidant activity. Z. Naturforsch. C 2000, 55, 1018–1022. [Google Scholar] [CrossRef] [PubMed]
- Huynh, B.L.C.; Duong, T.H.; Do, T.M.L.; Pinnock, T.G.; Pratt, L.M.; Yamamoto, S.; Watarai, H.; Tanahashi, T.; Nguyen, K.P.P. New gamma-Lactone Carboxylic Acids from the Lichen Parmotrema praesorediosum (Nyl.) Hale, Parmeliaceae. Rec. Nat. Prod. 2016, 10, 332–340. [Google Scholar]
- Huynh, B.L.C.; Le, D.H.; Takenaka, Y.; Tanahashi, T.; Nguyen, K.P.P. New phenolic compounds from the lichen Parmotrema praesorediosum (Nyl.) Hale (Parmeliaceae). Magn. Resonan. Chem. 2016, 54, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Tanabe, H.; Fujii-Kuriyama, Y.; Hayashi, H.; Inoue, M. Atranorin and lecanoric acid antagonize TCDD-induced xenobiotic response element-driven activity, but not xenobiotic response element-independent activity. J. Nat. Med. 2016, 70, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Holzmann, G.; Leuckert, C. Applications of negative fast atom bombardment and MS/MS to screening of lichen compounds. Phytochemistry 1990, 29, 2277–2283. [Google Scholar] [CrossRef]
- Parrot, D.; Jan, S.; Baert, N.; Guyot, S.; Tomasi, S. Comparative metabolite profiling and chemical study of Ramalina siliquosa complex using LC–ESI-MS/MS approach. Phytochemistry 2013, 89, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Parrot, D.; Peresse, T.; Hitti, E.; Carrie, D.; Grube, M.; Tomasi, S. Qualitative and spatial metabolite profiling of lichens by a LC-MS approach combined with optimised extraction. Phytochem. Anal. 2015, 26, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Le Pogam, P.; Schinkovitz, A.; Legouin, B.; Le Lamer, A.C.; Boustie, J.; Richomme, P. Matrix-Free UV-laser desorption ionization mass spectrometry as a versatile approach for accelerating dereplication studies on Lichens. Anal. Chem. 2015, 87, 10421–10428. [Google Scholar] [CrossRef] [PubMed]
- Musharraf, S.G.; Kanwal, N.; Thadhani, V.M.; Choudhary, M.I. Rapid identification of lichen compounds based on the structure-fragmentation relationship using ESI-MS/MS analysis. Anal. Methods 2015, 7, 6066–6076. [Google Scholar] [CrossRef]
- Simirgiotis, M.J.; Quispe, C.; Areche, C.; Sepulveda, B. Phenolic Compounds in Chilean Mistletoe (Quintral, Tristerix tetrandus) Analyzed by UHPLC-Q/Orbitrap/MS/MS and Its Antioxidant Properties. Molecules 2016, 21, 245. [Google Scholar] [CrossRef] [PubMed]
- Cornejo, A.; Salgado, F.; Caballero, J.; Vargas, R.; Simirgiotis, M.; Areche, C. Secondary metabolites in Ramalina terebrata detected by UHPLC/ESI/MS/MS and identification of parietin as Tau protein inhibitor. Int. J. Mol. Sci. 2016, 17, 1703. [Google Scholar] [CrossRef] [PubMed]
- Castro, O.N.; Benites, J.; Rodilla, J.; Santiago, J.C.; Simirgiotis, M.; Sepulveda, B.; Areche, C. Metabolomic Analysis of the Lichen Everniopsis trulla Using Ultra High Performance Liquid Chromatography-Quadrupole-Orbitrap Mass Spectrometry (UHPLC-Q-OT-MS). Chromatographia 2017, 80, 967–973. [Google Scholar] [CrossRef]
- Nobrega, N.D.; Ribeiro, S.M.; Pereira, E.C.; Marcelli, M.; Martins, M.C.B.; Falcao, E.P.D.; de Gusmao, N.B.; da Silva, N.H. Production of phenolic compounds from immobilized cells of the lichen Pamotrema andinum (Mull. Arg.) Hale and evaluation of antimicrobial activity. Acta Bot. Bras. 2012, 26, 101–107. [Google Scholar]
- Simirgiotis, M.J.; Ramirez, J.E.; Hirschmann, G.S.; Kennelly, E.J. Bioactive coumarins and HPLC-PDA-ESI-ToF-MS metabolic profiling of edible queule fruits (Gomortega keule), an endangered endemic Chilean species. Food Res. Int. 2013, 54, 532–543. [Google Scholar] [CrossRef]
- Bellio, P.; Di Pietro, L.; Mancini, A.; Piovano, M.; Nicoletti, M.; Brisdelli, F.; Tondi, D.; Cendron, L.; Franceschini, N.; Amicosante, G.; Perilli, M.; Celenza, G. SOS response in bacteria: Inhibitory activity of lichen secondary metabolites against Escherichia coli RecA protein. Phytomedicine 2017, 29, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.G.; Perez, E.M.R.; Padron, C.E.H.; Barrera, J.B. Chemical constituents of the lichen Stereocaulon azoreum. Z. Naturforsch. C 1992, 47, 503–507. [Google Scholar]
- Millanes, A.M.; Fontaniella, B.; Legaz, M.E.; Vicente, C. Histochemical detection of an haematommoyl alcohol dehydrogenase in the lichen Evernia prunastri. Plant Physiol. Biochem. 2003, 41, 786–791. [Google Scholar] [CrossRef]
- Soviar, K.; Motl, O.; Samek, Z.; Smolíková, J. The structure of lepraric acid, a lichen chromone. Tetrahedron Lett. 1967, 8, 2277–2279. [Google Scholar] [CrossRef]
- Latkowska, E.; Bober, B.; Chrapusta, E.; Adamski, M.; Kaminski, A.; Bialczyk, J. Secondary metabolites of the lichen Hypogymnia physodes (L.) Nyl. and their presence in spruce (Picea abies (L) H. Karst.) bark. Phytochemistry 2015, 118, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Gaikwad, S.; Verma, N.; Sharma, B.O.; Behera, B.C. Growth promoting effects of some lichen metabolites on probiotic bacteria. J. Food Sci. Technol. 2014, 51, 2624–2631. [Google Scholar] [CrossRef] [PubMed]
- Sutjaritturakan, J.; Saipunkaew, W.; Boonpragob, K.; Kalb, K. New species of Graphidaceae (Ostropales, Lecanoromycetes) from southern Thailand. Phytotaxa 2014, 189, 312–324. [Google Scholar] [CrossRef]
- Zlatanovic, I.; Zrnzevic, I.; Jovanovic, O.; Stojanovic, I.; Petrovic, G.; Stojanovic, G. Chemical Composition of Umbilicaria crustulosa and U. cylindrica. Nat. Prod. Commun. 2017, 12, 1105–1106. [Google Scholar]
Sample Availability: Samples of the lichens and some pure isolated compounds (Compound 50 = Usnic acid) are available from the authors. |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).