Identification of Anthocyanin Composition and Functional Analysis of an Anthocyanin Activator in Solanum nigrum Fruits
Abstract
:1. Introduction
2. Results
2.1. Analyses of Anthocyanins and their Antioxidant Activity in S. nigrum Fruits
2.2. Purification of Anthocyanins from S. nigrum Fruits
2.3. Identification of Anthocyanin Composition by Mass Spectrometry
2.4. Cloning of SnMYB from S. nigrum
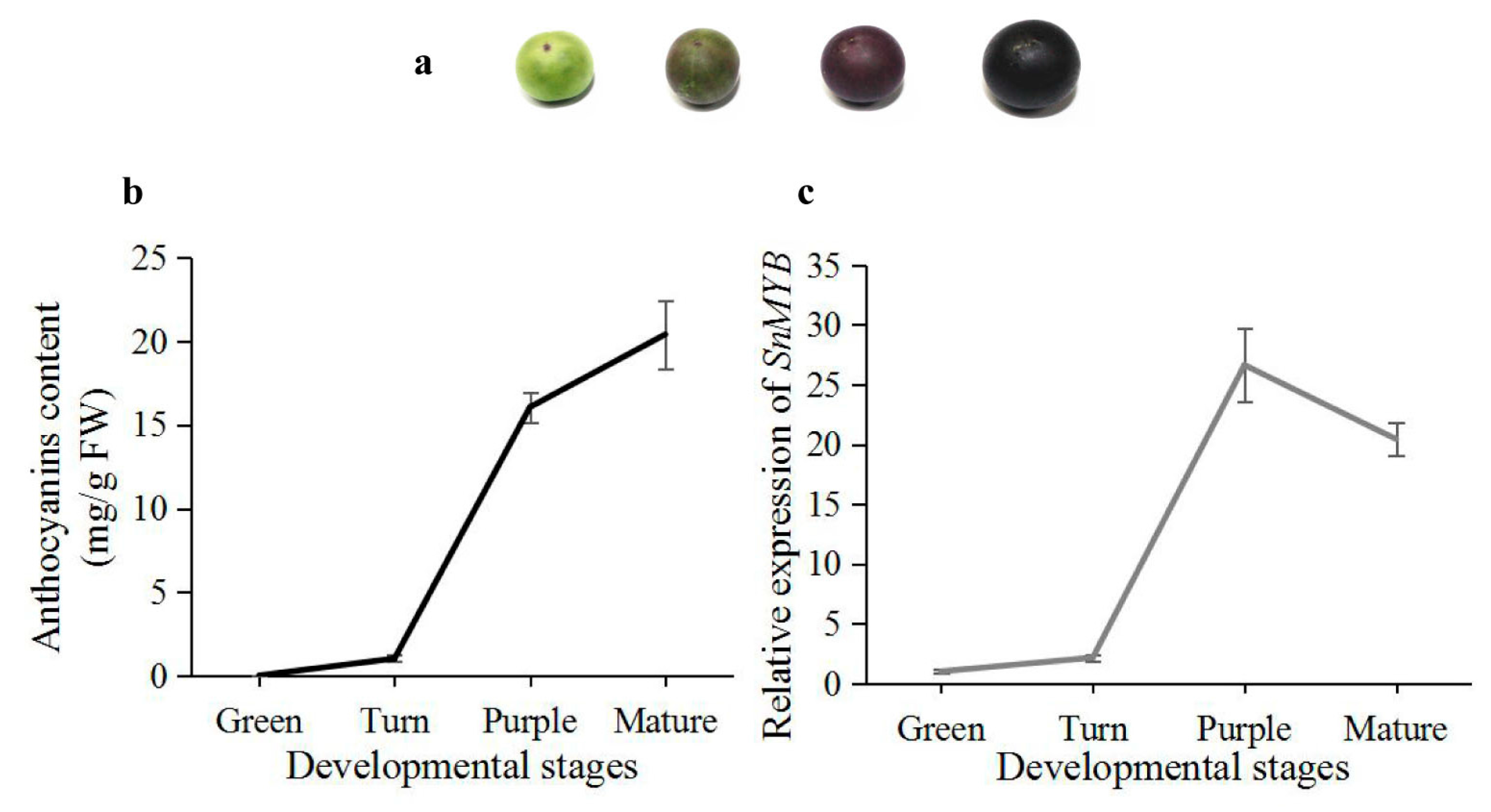
2.5. Expression Analysis of SnMYB in Different Developmental Stages of S. nigrum Fruits
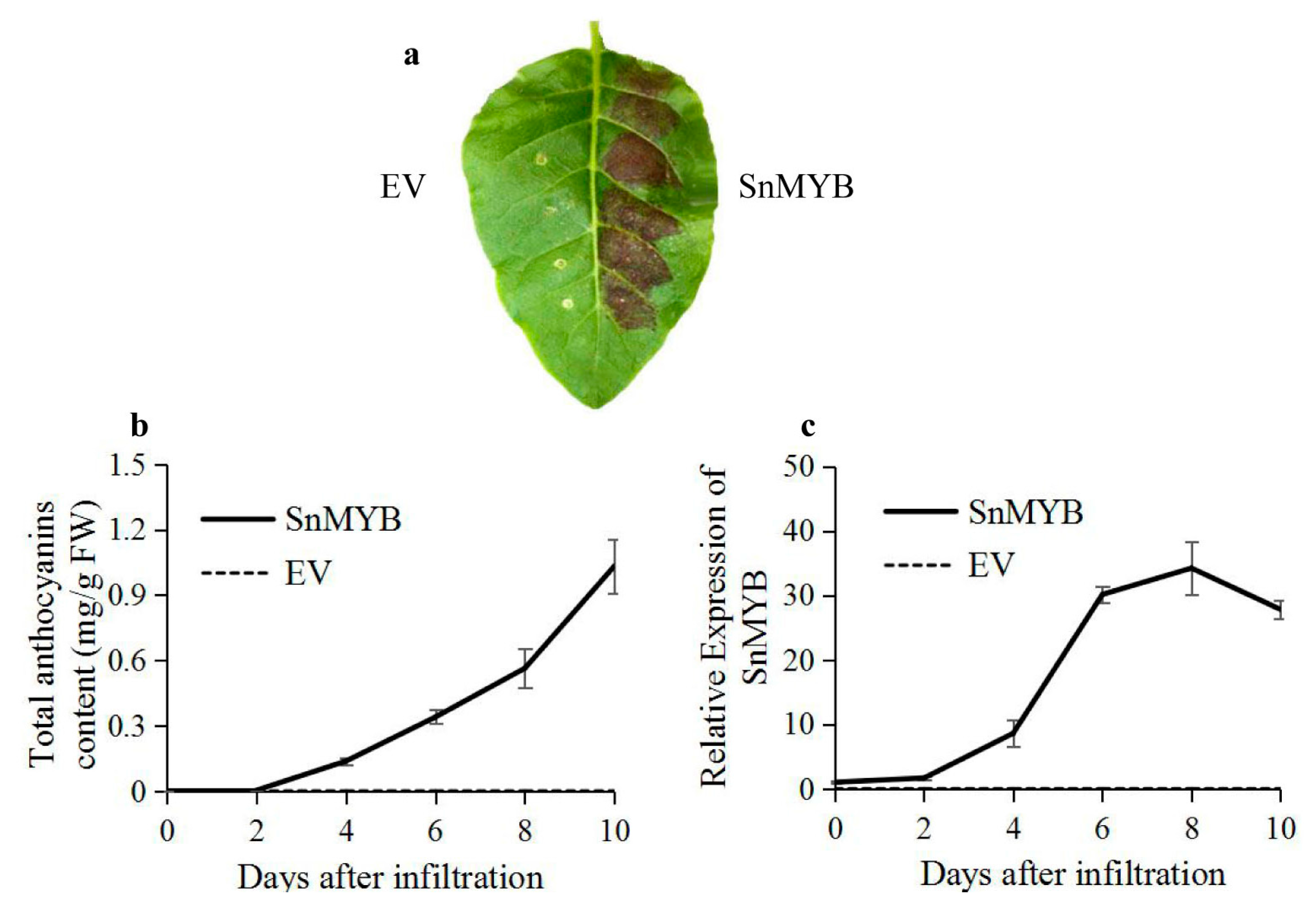
2.6. Transient Expression of SnMYB in Tobacco Leaves
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. HPLC and Mass Spectrometric Identification of Anthocyanins
4.3. Purification of Anthocyanins from S. nigrum Fruits
4.4. Purity Detection of Purified Anthocyanin Extracts
4.5. Total Antioxidant Activity Assay
4.6. RNA Extraction and Quantitative Real Time PCR
4.7. SnMYB Gene Cloning and Protein Sequence Alignment
4.8. Transient Assays of SnMYB Function
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Shang, Y.; Venail, J.; Mackay, S.; Bailey, P.C.; Schwinn, K.E.; Jameson, P.E.; Martin, C.R.; Davies, K.M. The molecular basis for venation patterning of pigmentation and its effect on pollinator attraction in flowers of Antirrhinum. New Phytol. 2011, 189, 602–615. [Google Scholar] [CrossRef] [PubMed]
- Ballaré, C.L. Stress under the sun: Spotlight on ultraviolet-B responses. Plant Physiol. 2003, 132, 1725–1727. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Butelli, E.; de Stefano, R.; Schoonbeek, H.J.; Magusin, A.; Pagliarani, C.; Wellner, N.; Hill, L.; Orzaez, D.; Granell, A.; et al. Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Curr. Biol. 2013, 23, 1094–1100. [Google Scholar] [CrossRef] [PubMed]
- Karageorgou, P.; Manetas, Y. The importance of being red when young: Anthocyanins and the protection of young leaves of Quercus coccifera from insect herbivory and excess light. Tree Physiol. 2006, 26, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Sui, Y.H.; Zhang, Z.X.; Xing, S.Z.; Lu, X.M.; Guo, Z.M. Effect of temperature on some physiologic indexes and germination of purple chili line YN 99007. Seed 2005, 24, 19–23. [Google Scholar]
- Tsuda, T.; Horio, F.; Uchida, K.; Aoki, H.; Osawa, T. Dietary cyanidin 3-O-β-d-glucoside-rich purple corn color prevents obesity and ameliorates hyperglycemia in mice. J. Nutr. 2003, 133, 2125–2130. [Google Scholar] [PubMed]
- Wang, L.S.; Kuo, C.T.; Cho, S.J.; Seguin, C.; Siddiqui, J.; Stoner, K.; Weng, Y.I.; Huang, T.H.; Tichelaar, J.; Yearsley, M.; et al. Black raspberry-derived anthocyanins demethylate tumor suppressor genes through the inhibition of DNMT1 and DNMT3B in colon cancer cells. Nutr. Cancer 2013, 65, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.G.; Yan, Q.Q.; Lu, L.Z.; Zhang, Y.Q. In vivo antioxidant, hypoglycemic, and anti-tumor activities of anthocyanin extracts from purple sweet potato. Nutr. Res. Pract. 2013, 7, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Kalt, W.; McDonald, J.E.; Fillmore, S.A.; Tremblay, F. Blueberry Effects on Dark Vision and Recovery after Photobleaching: Placebo-Controlled Crossover Studies. J. Agric. Food Chem. 2014, 62, 11180–11189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xu, X.; Zhang, J. Nutrition ingredient and exploitation of Solanum nigrum L. Chin. Wild Plant Resour. 2003, 23, 44–46. [Google Scholar]
- Zakaria, Z.A.; Gopalan, H.K.; Zainal, H.; Mohd Pojan, N.H.; Morsid, N.A.; Aris, A.; Sulaiman, M.R. Antinociceptive, anti-inflammatory and antipyretic effects of Solanum nigrum chloroform extract in animal models. Yakugaku Zasshi 2006, 126, 1171–1178. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.; Song, C.; Jin, C.A.; Qi, J.S. Determination and Analysis of Nutritional Ingredient in Solanum nigrum Fruit. Spec. Wild Econ. Anim. Plant Res. 2013, 2, 019. [Google Scholar]
- Huang, H.C.; Syu, K.Y.; Lin, J.K. Chemical composition of Solanum nigrum linn extract and induction of autophagy by leaf water extract and its major flavonoids in AU565 breast cancer cells. J. Agric. Food Chem. 2010, 58, 8699–8708. [Google Scholar] [CrossRef] [PubMed]
- Perez, G.R.M.; Perez, L.J.A.; Garcia, D.L.M.; Sossa, M.H. Neuropharmacological activity of Solanum nigrum fruit. J. Ethnopharmacol. 1998, 62, 43–48. [Google Scholar] [CrossRef]
- Sun, Y.; Zhou, Q.; Diao, C. Effects of cadmium and arsenic on growth and metal accumulation of Cd-hyperaccumulator Solanum nigrum L. Bioresour. Technol. 2008, 99, 1103–1110. [Google Scholar] [CrossRef] [PubMed]
- Strathearn, K.E.; Yousef, G.G.; Grace, M.H.; Roy, S.L.; Tambe, M.A.; Ferruzzi, M.G.; Wu, Q.M.; Simon, J.E.; Lila, M.A.; Rochet, J.C. Neuroprotective effects of anthocyanin-and proanthocyanidin-rich extracts in cellular models of Parkinson’s disease. Brain Res. 2014, 1555, 60–77. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Song, X.; Zhang, D.; Zhou, F.; Wang, D.; Wei, Y.; Gao, F.; Xie, L.; Jia, G.; Wu, W.; et al. Blueberry anthocyanins: Protection against ageing and light-induced damage in retinal pigment epithelial cells. Br. J. Nutr. 2012, 108, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Hichri, I.; Barrieu, F.; Bogs, J.; Kappel, C.; Delrot, S.; Lauvergeat, V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011, 62, 2465–2483. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Butelli, E.; Alseekh, S.; Tohge, T.; Rallapalli, G.; Luo, J.; Kawar, P.G.; Hill, L.; Santino, A.; Fernie, A.R.; et al. Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun. 2015, 6, 8635. [Google Scholar] [CrossRef] [PubMed]
- Fraser, L.G.; Seal, A.G.; Montefiori, M.; McGhie, T.K.; Tsang, G.K.; Datson, P.M.; Hilario, E.; Marsh, H.E.; Dunn, J.K.; Hellens, R.P.; et al. An R2R3 MYB transcription factor determines red petal colour in an Actinidia (kiwifruit) hybrid population. BMC Genom. 2013, 14, 28. [Google Scholar] [CrossRef] [PubMed]
- Pattanaik, S.; Kong, Q.; Zaitlin, D.; Werkman, J.R.; Xie, C.H.; Patra, B.; Yuan, L. Isolation and functional characterization of a floral tissue-specific R2R3 MYB regulator from tobacco. Planta 2010, 231, 1061–1076. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lin-Wang, K.; Espley, R.V.; Wang, L.; Yang, H.; Yu, B.; Dare, A.; Varkonyi-Gasic, E.; Wang, J.; Zhang, J.; et al. Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 2016, 67, 2159–2176. [Google Scholar] [CrossRef] [PubMed]
- Yuri, J.A.; Neira, A.; Quilodran, A.; Motomura, Y.; Palomo, I. Antioxidant activity and total phenolics concentration in apple peel and flesh is determined by cultivar and agroclimatic growing regions in Chile. J. Food Agric. Environ. 2009, 7, 3–4. [Google Scholar]
- Sellappan, S.; Akoh, C.C.; Krewer, G. Phenolic compounds and antioxidant capacity of Georgia-grown blueberries and blackberries. J. Agric. Food Chem. 2002, 50, 2432–2438. [Google Scholar] [CrossRef] [PubMed]
- Prior, R.L.; Cao, G.; Martin, A.; Sofic, E.; McEwen, J.; O'Brien, C.; Lischner, N.; Ehlenfeldt, M.; Kalt, W.; Krewer, G.; et al. Antioxidant capacity as influenced by total phenolic and anthocyanin content, maturity, and variety of Vaccinium species. J. Agric. Food Chem. 1998, 46, 2686–2693. [Google Scholar] [CrossRef]
- Moyer, R.A.; Hummer, K.E.; Finn, C.E.; Frei, B.; Wrolstad, R.E. Anthocyanins, phenolics, and antioxidant capacity in diverse small fruits: Vaccinium, Rubus, and Ribes. J. Agric. Food Chem. 2002, 50, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Oszmianski, J.; Kolniak-Ostek, J.; Wojdyło, A. Characterization of phenolic compounds and antioxidant activity of Solanum scabrum and Solanum burbankii berries. J. Agric. Food Chem. 2014, 62, 1512–1519. [Google Scholar] [CrossRef] [PubMed]
- Kirakosyan, A.; Seymour, E.M.; Wolforth, J.; McNish, R.; Kaufman, P.B.; Bolling, S.F. Tissue bioavailability of anthocyanins from whole tart cherry in healthy rats. Food Chem. 2015, 171, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Ding, C.; Wang, L.; Li, G.; Shi, J.; Li, H.; Wang, H.; Suo, Y. Anthocyanins composition and antioxidant activity of wild Lycium ruthenicum Murr. from Qinghai-Tibet Plateau. Food Chem. 2011, 126, 859–865. [Google Scholar] [CrossRef]
- Su, X.; Xu, J.; Rhodes, D.; Shen, Y.; Song, W.; Katz, B.; Tomich, J.; Wang, W. Identification and quantification of anthocyanins in transgenic purple tomato. Food Chem. 2016, 202, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Wu, B.; Fan, P.; Yang, C.; Duan, W.; Zheng, X.; Liu, C.; Li, S. Anthocyanin composition and content in grape berry skin in Vitis germplasm. Food Chem. 2008, 111, 837–844. [Google Scholar] [CrossRef]
- Zimmermann, I.M.; Heim, M.A.; Weisshaar, B.; Uhrig, J.F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004, 40, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Lin-Wang, K.; Bolitho, K.; Grafton, K.; Kortstee, A.; Karunairetnam, S.; McGhie, T.K.; Espley, R.V.; Hellens, R.P.; Allan, A.C. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010, 10. [Google Scholar] [CrossRef] [PubMed]
- Hichri, I.; Heppel, S.C.; Pillet, J.; Léon, C.; Czemmel, S.; Delrot, S.; Lauvergeat, V.; Bogs, J. The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol. Plant 2010, 3, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Gavrilova, V.; Kajdzanoska, M.; Gjamovski, V.; Stefova, M. Separation, Characterization and Quantification of Phenolic Compounds in Blueberries and Red and Black Currants by HPLC-DAD-ESI-MSn. J. Agric. Food Chem. 2011, 59, 4009–4018. [Google Scholar] [CrossRef] [PubMed]
- Drogoudi, P.D.; Michailidis, Z.; Pantelidis, G. Peel and flesh antioxidant content and harvest quality characteristics of seven apple cultivars. Sci. Hortic. 2008, 115, 149–153. [Google Scholar] [CrossRef]
- Lapornik, B.; Prošek, M.; Wondra, A.G. Comparison of extracts prepared from plant by-products using different solvents and extraction time. J. Food Eng. 2005, 71, 214–222. [Google Scholar] [CrossRef]
- Ban, Y.; Honda, C.; Hatsuyama, Y.; Igarashi, M.; Bessho, H.; Moriguchi, T. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 2007, 48, 958–970. [Google Scholar] [CrossRef] [PubMed]
- Montefiori, M.; Brendolise, C.; Dare, A.P.; Lin-Wang, K.; Davies, K.M.; Hellens, R.P.; Allan, A.C. In the Solanaceae, a hierarchy of bHLHs confer distinct target specificity to the anthocyanin regulatory complex. J. Exp. Bot. 2015, 66, 1427–1436. [Google Scholar] [CrossRef] [PubMed]
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.P.; Matros, A.; Peterek, S.; Schijlen, E.G.W.M.; Hall, R.D.; Bovy, A.G.; Luo, J.; et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 2008, 26, 1301–1308. [Google Scholar] [CrossRef] [PubMed]
- Chandra, A.; Rana, J.; Li, Y. Separation, identification, quantification, and method validation of anthocyanins in botanical supplement raw materials by HPLC and HPLC-MS. J. Agric. Food Chem. 2001, 49, 3515–3521. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Yin, Y.; Xu, C.; Liu, J. Isolation of high-purity anthocyanin mixtures and monomers from blueberries using combined chromatographic techniques. J. Chromatogr. A 2014, 1327, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tang, W.; Chen, J.; Jia, R.; Ma, L.; Wang, S.; Wang, J.; Shen, X.; Chu, Z.; Zhu, C.; et al. Development of marker-free transgenic potato tubers enriched in caffeoylquinic acids and flavonols. J. Agric. Food Chem. 2016, 64, 2932–2940. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Malcuit, I.; Moffett, P.; Ruiz, M.T.; Peart, J.; Wu, A.J.; Rathjen, J.P.; Bendahmane, A.; Day, L.; Baulcombe, D.C. High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 2003, 22, 5690–5699. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.H.; Sohn, S.H.; Kim, D.H.; Kim, J.K.; Lee, J.Y.; Kim, Y.M.; Ha, S.H. Use of an anthocyanin production phenotype as a visible selection marker system in transgenic tobacco plant. Plant Biotechnol. Rep. 2012, 6, 203–211. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compound petunidin-3-(trans-coumaroyl)-rutinoside-5-glucoside is available from the authors. |

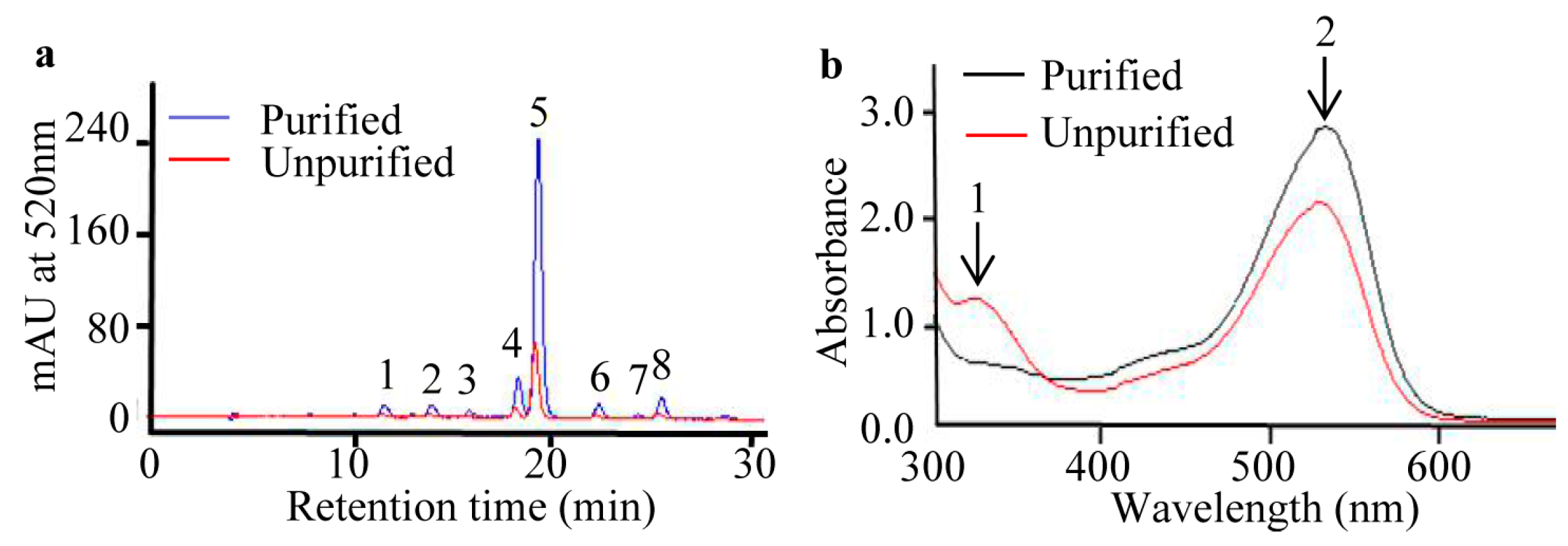
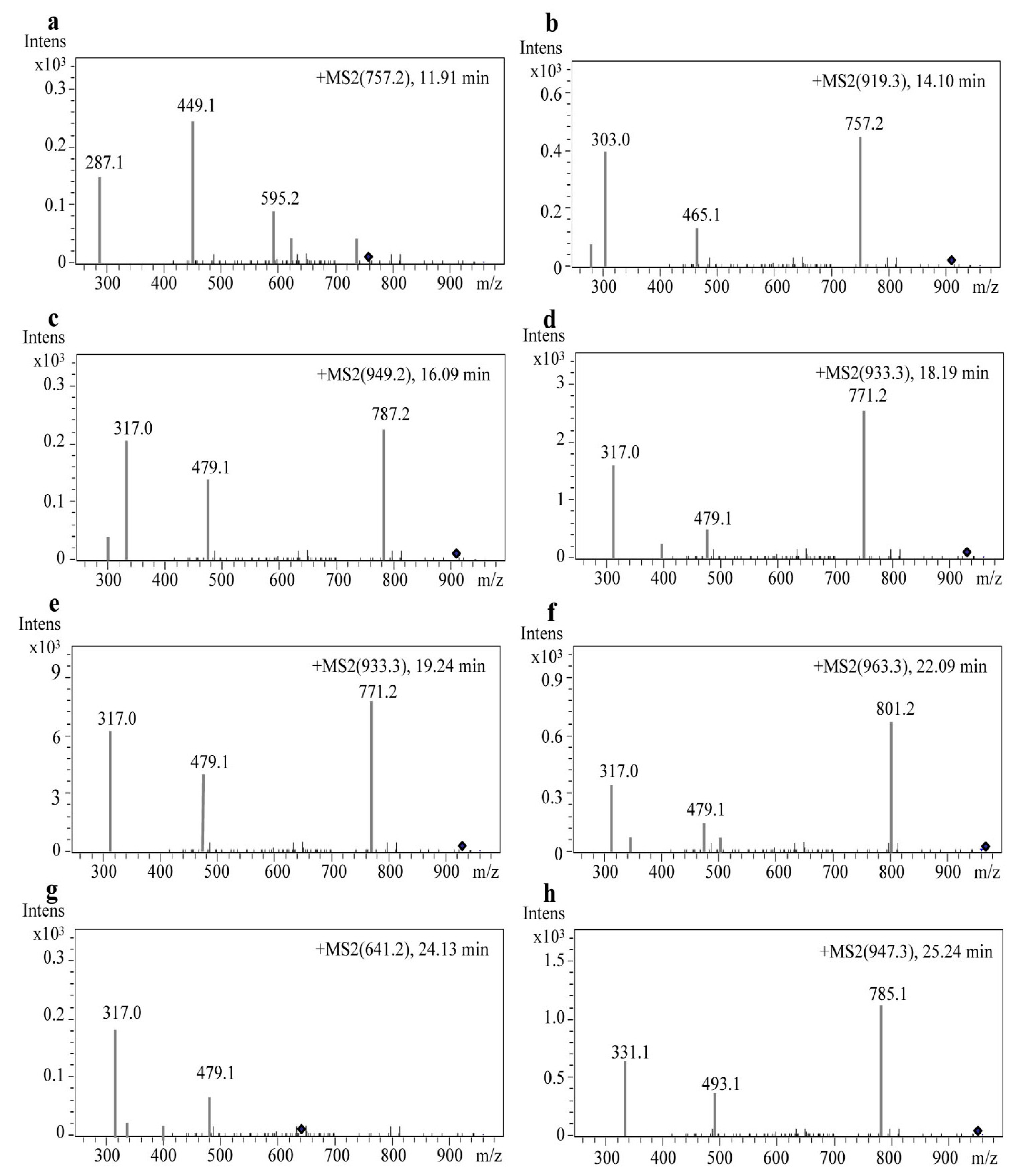

| Peak No. | Retention Time (min) | Anthocyanins | [M + H]+ (m/z) | Detected Fragments |
|---|---|---|---|---|
| 1 | 11.921 | Cyanidin-3-rutinoside-5-glucoside | 757.2 | 595.22; 449.15; 287.08 |
| 2 | 14.109 | Delphinidin-3-(p-coumaroyl)-rutinoside-5-glucoside | 919.3521 | 757.27; 465.15; 303.08 |
| 3 | 16.093 | Petunidin-3-O-rutinoside-(caffeoyl)-5-O-glucoside | 949.2482 | 787.26; 479.15; 317.08 |
| 4 | 18.191 | Petunidin-3-(cis-p-coumaroyl)-rutinoside-5-glucoside | 933.3681 | 771.29; 479.16; 317.09 |
| 5 | 19.240 | Petunidin-3-(trans-p-coumaroyl)-rutinoside-5-glucoside | 933.3677 | 771.29; 479.17; 317.09 |
| 6 | 22.093 | Petunidin-3-(feruloyl)-rutinoside-5-glucoside | 963.3784 | 801.30; 479.16; 317.09 |
| 7 | 24.139 | Petunidin-3-O-glucoside-5-O-glucoside | 641.2 | 479.16; 317.09 |
| 8 | 25.242 | Malvidin-3-(p-coumaroyl)-rutinoside-5-glucoside | 947.3831 | 785.31; 493.18; 331.11 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Chu, Z.; Ren, M.; Jia, R.; Zhao, C.; Fei, D.; Su, H.; Fan, X.; Zhang, X.; Li, Y.; et al. Identification of Anthocyanin Composition and Functional Analysis of an Anthocyanin Activator in Solanum nigrum Fruits. Molecules 2017, 22, 876. https://doi.org/10.3390/molecules22060876
Wang S, Chu Z, Ren M, Jia R, Zhao C, Fei D, Su H, Fan X, Zhang X, Li Y, et al. Identification of Anthocyanin Composition and Functional Analysis of an Anthocyanin Activator in Solanum nigrum Fruits. Molecules. 2017; 22(6):876. https://doi.org/10.3390/molecules22060876
Chicago/Turabian StyleWang, Shaoli, Zhaohui Chu, Mingxing Ren, Ru Jia, Changbao Zhao, Dan Fei, Hao Su, Xiaoqi Fan, Xiaotian Zhang, Yang Li, and et al. 2017. "Identification of Anthocyanin Composition and Functional Analysis of an Anthocyanin Activator in Solanum nigrum Fruits" Molecules 22, no. 6: 876. https://doi.org/10.3390/molecules22060876
APA StyleWang, S., Chu, Z., Ren, M., Jia, R., Zhao, C., Fei, D., Su, H., Fan, X., Zhang, X., Li, Y., Wang, Y., & Ding, X. (2017). Identification of Anthocyanin Composition and Functional Analysis of an Anthocyanin Activator in Solanum nigrum Fruits. Molecules, 22(6), 876. https://doi.org/10.3390/molecules22060876






