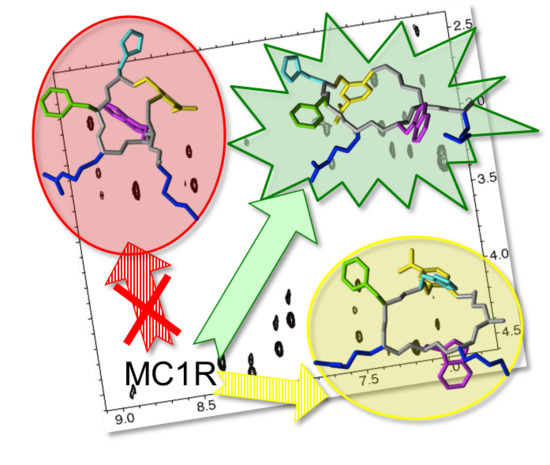
NMR Insights into the Structure-Function Relationships in the Binding of Melanocortin Analogues to the MC1R Receptor
Abstract
:1. Introduction
2. Results
2.1. Affinity of Peptides CycN-K6 and CycN-K7 to MCR1
2.2. NMR Characterization of CycN-K6 and CycN-K7 and Comparison to CycS-C6
2.3. NMR Solution Structures of Analogues CycN-K6 and CycN-K7
3. Discussion
4. Materials and Methods
4.1. Peptide Synthesis
4.2. Competitive Binding Assays
4.3. NMR Sample Preparation
4.4. Acquisition of NMR Spectra
4.5. NMR Spectral Assignment
4.6. Structure Calculation
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chhajlani, V.; Wikberg, J.E. Molecular cloning and expression of the human melanocyte stimulating hormone receptor cDNA. FEBS Lett. 1992, 309, 417–420. [Google Scholar] [CrossRef]
- Gerst, J.E.; Sole, J.; Hazum, E.; Salomon, Y. Identification and characterization of melanotropin binding proteins from M2R melanoma cells by covalent photoaffinity labeling. Endocrinology 1988, 123, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Muceniece, R.; Mutule, I.; Mutulis, F.; Prusis, P.; Szardenings, M.; Wikberg, J.E. Detection of regions in the MC1 receptor of importance for the selectivity of the MC1 receptor super-selective MS04/MS05 peptides. Biochim. Biophys. Acta 2001, 1544, 278–282. [Google Scholar] [CrossRef]
- Solca, F.; Siegrist, W.; Drozdz, R.; Girard, J.; Eberle, A.N. The receptor for alpha-melanotropin of mouse and human melanoma cells. Application of a potent alpha-melanotropin photoaffinity label. J. Biol. Chem. 1989, 264, 14277–14281. [Google Scholar] [PubMed]
- Yang, Y. Structure, function and regulation of the melanocortin receptors. Eur. J. Pharmacol. 2011, 660, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Mountjoy, K.G.; Robbins, L.S.; Mortrud, M.T.; Cone, R.D. The cloning of a family of genes that encode the melanocortin receptors. Science 1992, 257, 1248–1251. [Google Scholar] [CrossRef] [PubMed]
- Holder, J.R.; Haskell-Luevano, C. Melanocortin ligands: 30 years of structure-activity relationship (SAR) studies. Med. Res. Rev. 2004, 24, 325–356. [Google Scholar] [CrossRef] [PubMed]
- Van der Velden, P.A.; Sandkuijl, L.A.; Bergman, W.; Pavel, S.; Van Mourik, L.; Frants, R.R.; Gruis, N.A. Melanocortin-1 receptor variant R151C modifies melanoma risk in dutch families with melanoma. Am. J. Hum. Genet. 2001, 69, 774–779. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.C.; Wakamatsu, K.; Ito, S.; Kadekaro, A.L.; Kobayashi, N.; Groden, J.; Kavanagh, R.; Takakuwa, T.; Virador, V.; Hearing, V.J.; et al. Human melanocortin 1 receptor variants, receptor function and melanocyte response to UV radiation. J. Cell Sci. 2002, 115, 2349–2355. [Google Scholar] [PubMed]
- Siegrist, W.; Solca, F.; Stutz, S.; Giuffre, L.; Carrel, S.; Girard, J.; Eberle, A.N. Characterization of receptors for alpha-melanocyte-stimulating hormone on human melanoma cells. Cancer Res. 1989, 49, 6352–6358. [Google Scholar] [PubMed]
- Hruby, V.J.; Cai, M.; Cain, J.; Nyberg, J.; Trivedi, D. Design of novel melanocortin receptor ligands: Multiple receptors, complex pharmacology, the challenge. Eur. J. Pharmacol. 2011, 660, 88–93. [Google Scholar] [CrossRef] [PubMed]
- García-Borrón, J.C.; Sánchez-Laorden, B.L.; Jiménez-Cervantes, C. Melanocortin-1 receptor structure and functional regulation. Pigment Cell Res. 2005, 18, 393–410. [Google Scholar] [CrossRef] [PubMed]
- Raposinho, P.D.; Correia, J.D.; Oliveira, M.C.; Santos, I. Melanocortin-1 receptor-targeting with radiolabeled cyclic alpha-melanocyte-stimulating hormone analogs for melanoma imaging. Biopolymers 2010, 94, 820–829. [Google Scholar] [CrossRef] [PubMed]
- Correia, J.D.; Paulo, A.; Raposinho, P.D.; Santos, I. Radiometallated peptides for molecular imaging and targeted therapy. Dalton Trans. 2011, 40, 6144–6167. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Cheng, Z.; Hoffman, T.J.; Jurisson, S.S.; Quinn, T.P. Melanoma-targeting properties of (99 m) technetium-labeled cyclic alpha-melanocyte-stimulating hormone peptide analogues. Cancer Res. 2000, 60, 5649–5658. [Google Scholar] [PubMed]
- Li, Z.B.; Cai, W.; Cao, Q.; Chen, K.; Wu, Z.; He, L.; Chen, X. 64Cu-labeled tetrameric and octameric RGD peptides for small-animal pet of tumor alpha(v)beta(3) integrin expression. J. Nucl. Med. 2007, 48, 1162–1171. [Google Scholar] [CrossRef] [PubMed]
- Barkey, N.M.; Tafreshi, N.K.; Josan, J.S.; De Silva, C.R.; Sill, K.N.; Hruby, V.J.; Gillies, R.J.; Morse, D.L.; Vagner, J. Development of melanoma-targeted polymer micelles by conjugation of a melanocortin 1 receptor (MC1R) specific ligand. J. Med. Chem. 2011, 54, 8078–8084. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Sihver, W.; Jurischka, C.; Bergmann, R.; Haase-Kohn, C.; Mosch, B.; Steinbach, J.; Carta, D.; Bolzati, C.; Calderan, A.; et al. Radiopharmacological characterization of 64Cu-labeled alpha-MSH analogs for potential use in imaging of malignant melanoma. Amino Acids 2016, 48, 833–847. [Google Scholar] [CrossRef] [PubMed]
- Grieco, P.; Cai, M.; Liu, L.; Mayorov, A.; Chandler, K.; Trivedi, D.; Lin, G.; Campiglia, P.; Novellino, E.; Hruby, V.J. Design and microwave-assisted synthesis of novel macrocyclic peptides active at melanocortin receptors: Discovery of potent and selective hMC5R receptor antagonists. J. Med. Chem. 2008, 51, 2701–2707. [Google Scholar] [CrossRef] [PubMed]
- Morais, M.; Raposinho, P.D.; Oliveira, M.C.; Pantoja-Uceda, D.; Jimenez, M.A.; Santos, I.; Correia, J.D.G. NMR structural analysis of MC1R-targeted rhenium(i) metallopeptides and biological evaluation of Tc-99m(i) congeners. Organometallics 2012, 31, 5929–5939. [Google Scholar] [CrossRef]
- Wishart, D.S.; Bigam, C.G.; Holm, A.; Hodges, R.S.; Sykes, B.D. H-1, C-13 and N-15 random coil nmr chemical-shifts of the common amino-acids. 1. Investigations of nearest-neighbor effects. J. Biomol. NMR 1995, 5, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Merutka, G.; Dyson, H.J.; Wright, P.E. Random coil H-1 chemical-shifts obtained as a function of temperature and trifluoroethanol concentration for the peptide series ggxgg. J. Biomol. NMR 1995, 5, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, A.C.; Kondejewski, L.H.; Gronwald, W.; Nip, A.M.; Hodges, R.S.; Sykes, B.D.; Wishart, D.S. Unusual beta-sheet periodicity in small cyclic peptides. Nat. Struct. Biol. 1998, 5, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Prusis, P.; Schioth, H.B.; Muceniece, R.; Herzyk, P.; Afshar, M.; Hubbard, R.E.; Wikberg, J.E. Modeling of the three-dimensional structure of the human melanocortin 1 receptor, using an automated method and docking of a rigid cyclic melanocyte-stimulating hormone core peptide. J. Mol. Graph. Model. 1997, 15, 307–317, 334. [Google Scholar] [CrossRef]
- Prusis, P.; Frandberg, P.A.; Muceniece, R.; Kalvinsh, I.; Wikberg, J.E. A three dimensional model for the interaction of msh with the melanocortin-1 receptor. Biochem. Biophys. Res. Commun. 1995, 210, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Schioth, H.B.; Muceniece, R.; Szardenings, M.; Prusis, P.; Wikberg, J.E. Evidence indicating that the TM4, EL2, and TM5 of the melanocortin 3 receptor do not participate in ligand binding. Biochem. Biophys. Res. Commun. 1996, 229, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Falcao, C.B.; Pérez-Peinado, C.; de la Torre, B.G.; Mayol, X.; Zamora-Carreras, H.; Jiménez, M.A.; Radis-Baptista, G.; Andreu, D. Structural dissection of crotalicidin, a rattlesnake venom cathelicidin, retrieves a fragment with antimicrobial and antitumor activity. J. Med. Chem. 2015, 58, 8553–8563. [Google Scholar] [CrossRef] [PubMed]
- Baeza, J.L.; de la Torre, B.G.; Santiveri, C.M.; Almeida, R.D.; García-López, M.T.; Gerona-Navarro, G.; Jaffrey, S.R.; Jiménez, M.A.; Andreu, D.; González-Muñiz, R.; et al. Cyclic amino acid linkers stabilizing key loops of brain derived neurotrophic factor. Bioorg. Med. Chem. Lett. 2012, 22, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.K.; Lee, C.J.; Lee, C.H.; Li, S.Z.; Lim, S.K.; Baik, J.H.; Lee, W. Structure and function of the potent cyclic and linear melanocortin analogues. J. Struct. Biol. 2005, 150, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Mirassou, Y.; Santiveri, C.M.; Pérez de Vega, M.J.; González-Muñiz, R.; Jiménez, M.A. Disulfide bonds versus Trp Trp pairs in irregular beta-hairpins: NMR structure of vammin loop 3-derived peptides as a case study. ChemBioChem 2009, 10, 902–910. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Bigam, C.G.; Yao, J.; Abildgaard, F.; Dyson, H.J.; Oldfield, E.; Markley, J.L.; Sykes, B.D. H-1, C-13 and N-15 chemical-shift referencing in biomolecular NMR. J. Biomol. NMR 1995, 6, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Goddard, T.D.; Kneller, D.G. SPARKY 3. Available online: https://www.cgl.ucsf.edu/home/sparky/ (accessed on 1 December 2011).
- Wuthrich, K. NMR of Proteins and Nucleic Acids; John Wiley & Sons: New York, NY, USA, 1986. [Google Scholar]
- Wuthrich, K.; Billeter, M.; Braun, W. Polypeptide secondary structure determination by nuclear magnetic resonance observation of short proton-proton distances. J. Mol. Biol. 1984, 180, 715–740. [Google Scholar] [CrossRef]
- Guntert, P. Automated NMR structure calculation with cyana. Methods Mol. Biol. 2004, 278, 353–378. [Google Scholar] [PubMed]
- Guntert, P.; Mumenthaler, C.; Wuthrich, K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997, 273, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Koradi, R.; Billeter, M.; Wuthrich, K. Molmol: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996, 14, 51–55, 29–32. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |




| Peptide | Sequence | Cycle Size | IC50 (nM) |
|---|---|---|---|
| PG10N 1 | c[S–NO2–C6H3–CO–His–DPhe–Arg–Trp–Cys]–NH2 | 19 | 3.7 ± 0.5 |
| CycS-C6 1 | c[S–NO2–C6H3–CO–His–DPhe–Arg–Trp–Cys]–Lys–NH2 | 19 | 1770 ± 480 |
| CycN-K6 | c[NH–NO2–C6H3–CO–His–DPhe–Arg–Trp–Lys]–Lys–NH2 | 22 | 155 ± 16 |
| CycN-K7 | c[NH–NO2–C6H3–CO–His–DPhe–Arg–Trp–Lys–Lys]–NH2 | 25 | 495 ± 101 |
| CycS-C6 2 | CycN-K6 | CycN-K7 | Random Coil 3 | |
|---|---|---|---|---|
| Cycle size (number atoms) | 19 | 22 | 25 | |
| 3JαN His (Hz) 4 | 7.9 | 4.2 | 6.0 | 5.5–7.4 5 |
| 3JαN DPhe (Hz) 4 | 4.8 | 6.6 | 6.2 | 5.5–7.4 5 |
| δ Hα His (ppm) | 4.96 | 4.68 | 4.85 | 4.73 |
| δ Hα Arg (ppm) | 3.99 | 4.42 | 4.13 | 4.34 |
| δ Hββ’ Arg (ppm) | 1.12, 1.38 | 1.48, 1.69 | 1.41, 1.60 | 1.76, 1.86 |
| δ Hγγ’ Arg (ppm) | 0.59, 0.79 | 1.02, 1.02 | 0.94, 1.08 | 1.63, 1.63 |
| δ Hδδ’ Arg (ppm) | 2.78, 2.78 | 2.48, 2.82 | 2.71, 2.85 | 3.20, 3.20 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morais, M.; Zamora-Carreras, H.; Raposinho, P.D.; Oliveira, M.C.; Pantoja-Uceda, D.; Correia, J.D.G.; Jiménez, M.A. NMR Insights into the Structure-Function Relationships in the Binding of Melanocortin Analogues to the MC1R Receptor. Molecules 2017, 22, 1189. https://doi.org/10.3390/molecules22071189
Morais M, Zamora-Carreras H, Raposinho PD, Oliveira MC, Pantoja-Uceda D, Correia JDG, Jiménez MA. NMR Insights into the Structure-Function Relationships in the Binding of Melanocortin Analogues to the MC1R Receptor. Molecules. 2017; 22(7):1189. https://doi.org/10.3390/molecules22071189
Chicago/Turabian StyleMorais, Maurício, Héctor Zamora-Carreras, Paula D. Raposinho, Maria Cristina Oliveira, David Pantoja-Uceda, João D. G. Correia, and M. Angeles Jiménez. 2017. "NMR Insights into the Structure-Function Relationships in the Binding of Melanocortin Analogues to the MC1R Receptor" Molecules 22, no. 7: 1189. https://doi.org/10.3390/molecules22071189