Predicting and Interpreting the Structure of Type IV Pilus of Electricigens by Molecular Dynamics Simulations
Abstract
:1. Introduction
2. Results and Discussion
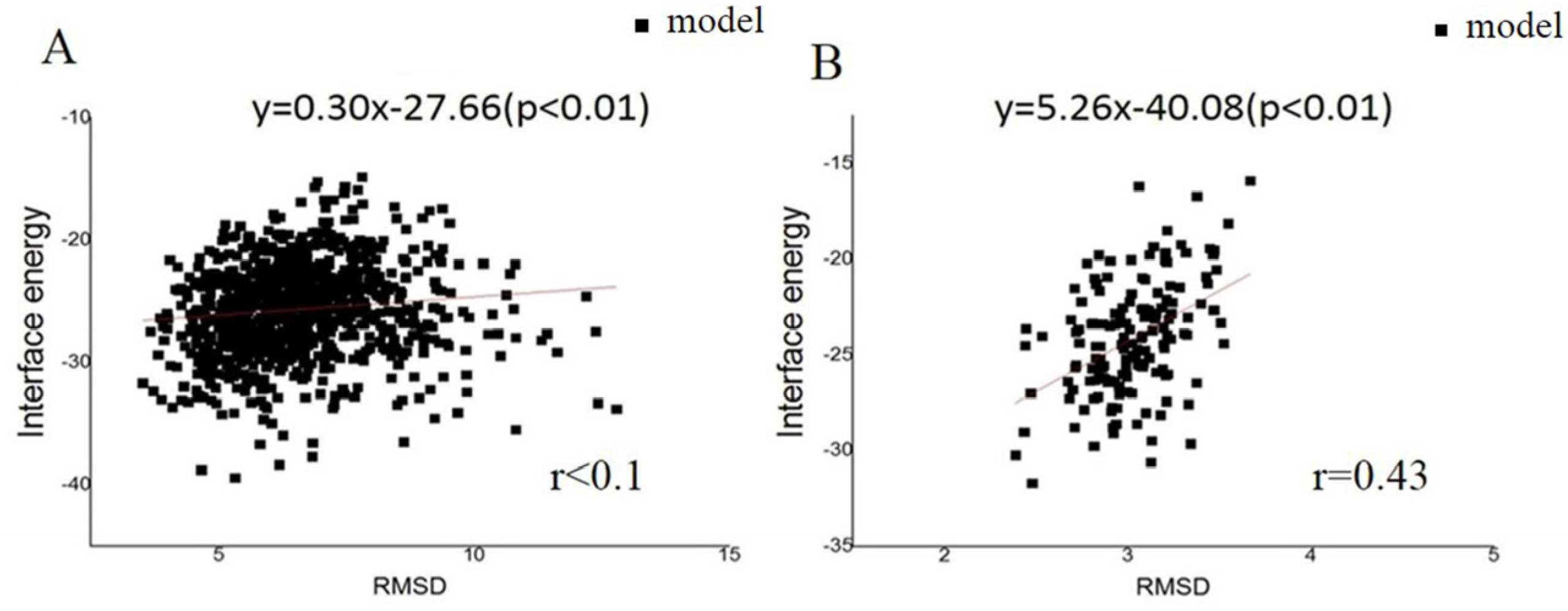
2.1. Predicted GC Pilus Was Closer to the Native Structure Than Previous Model
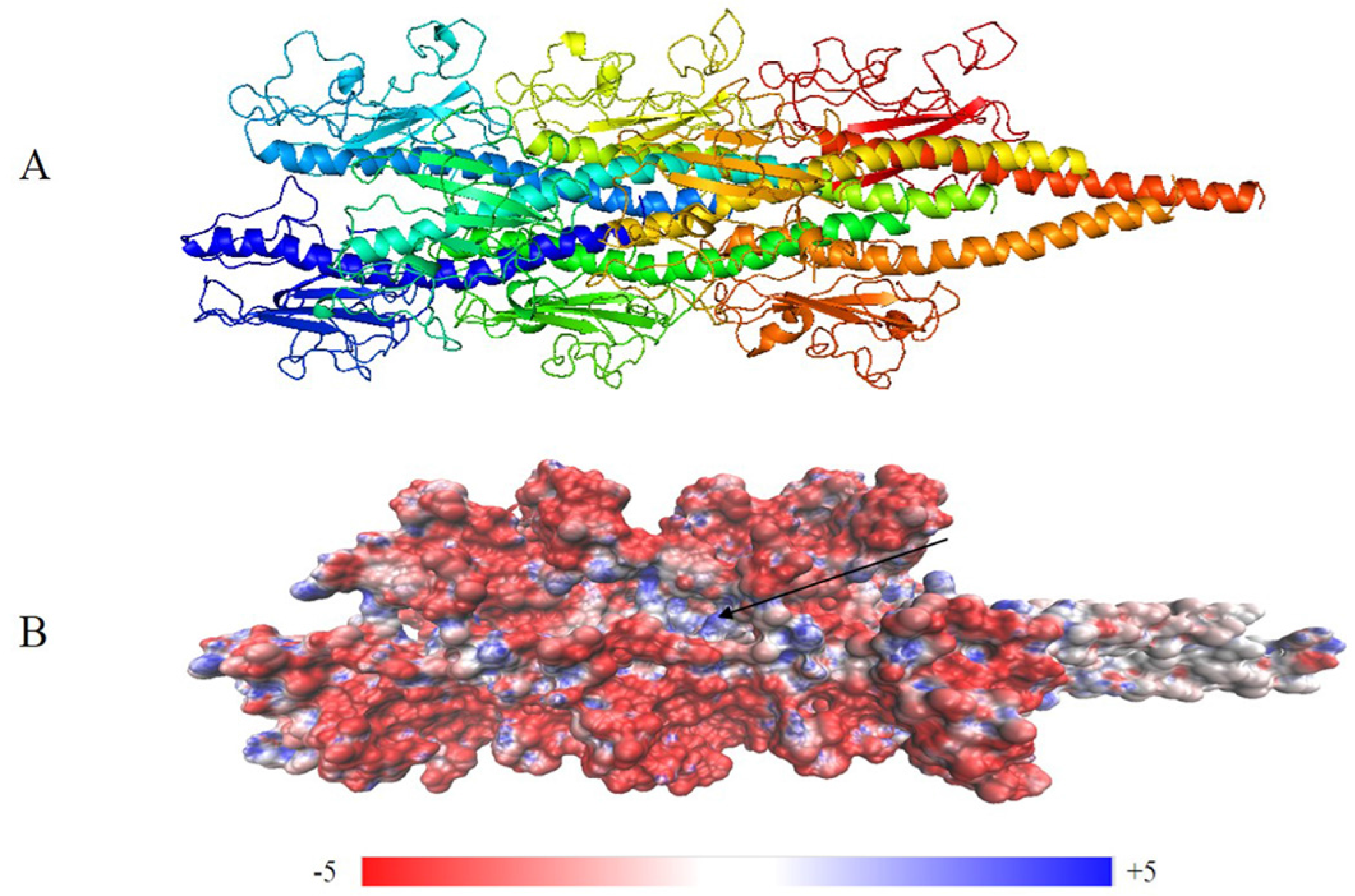
2.2. Structure of the G. Uraniireducens Pilus
2.2.1. Monomer Structure of the G. uraniireducens Pilus
2.2.2. Predicted Structure of G. uraniireducens Pilus
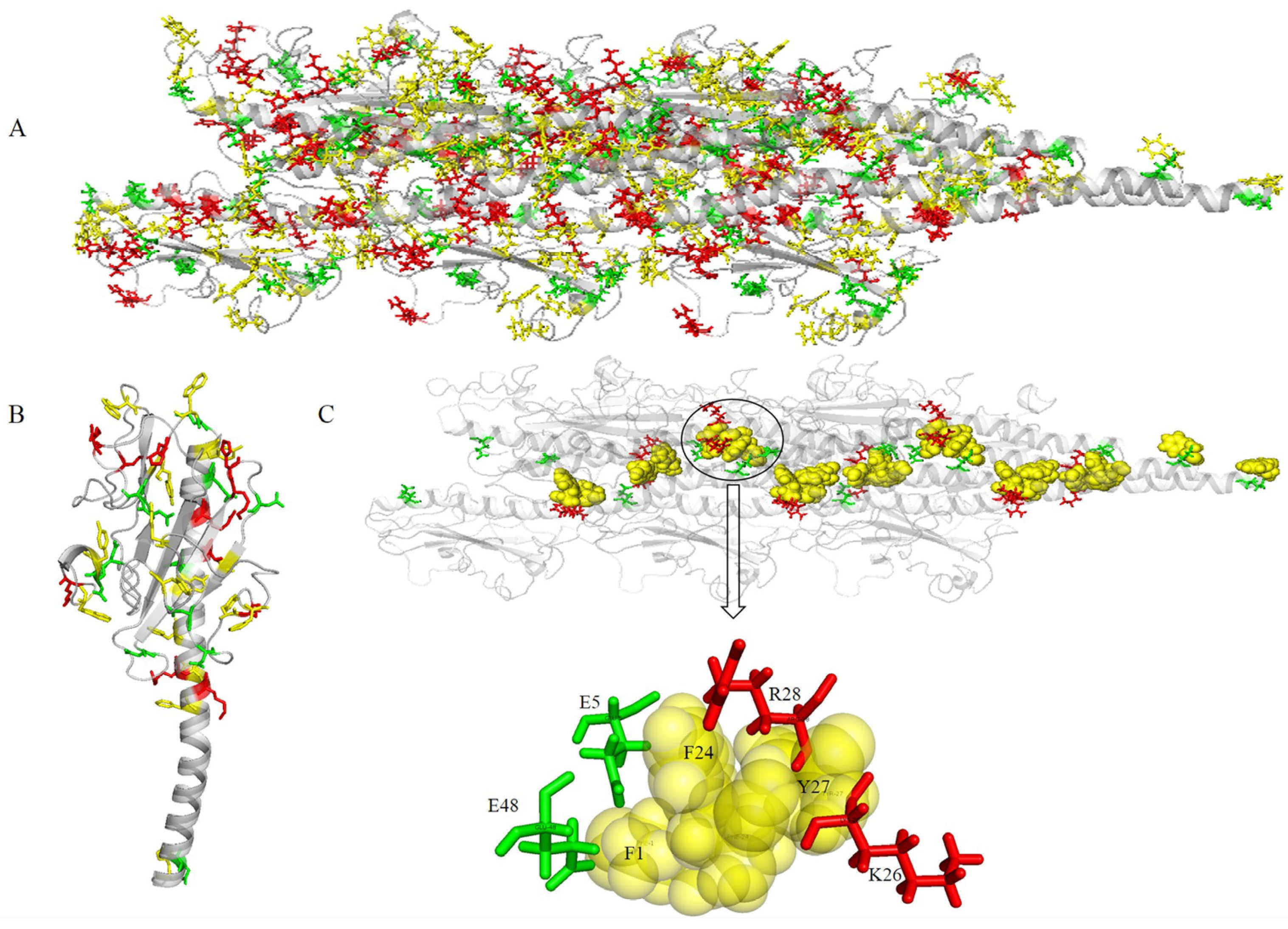
2.3. Potential Structural Basis for Conductivity in G. uraniireducens Pili
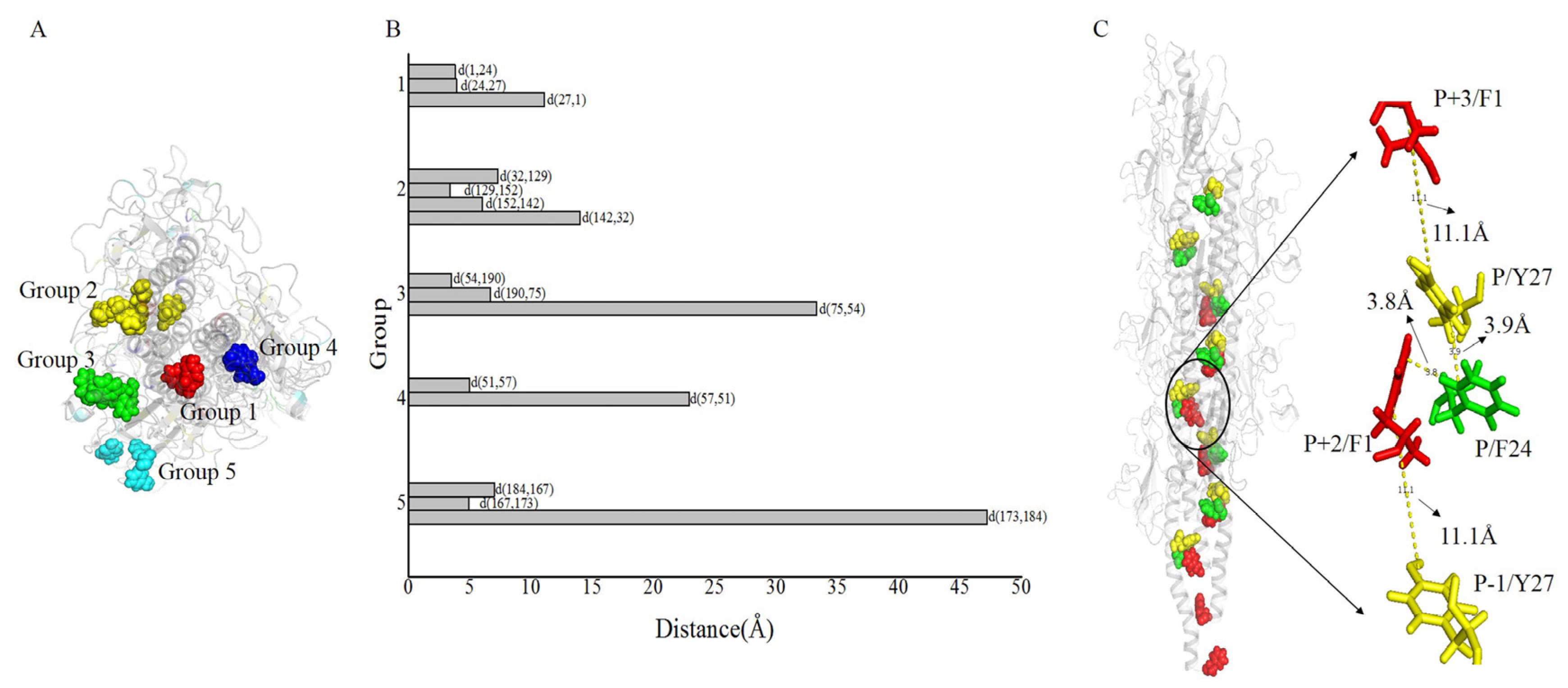
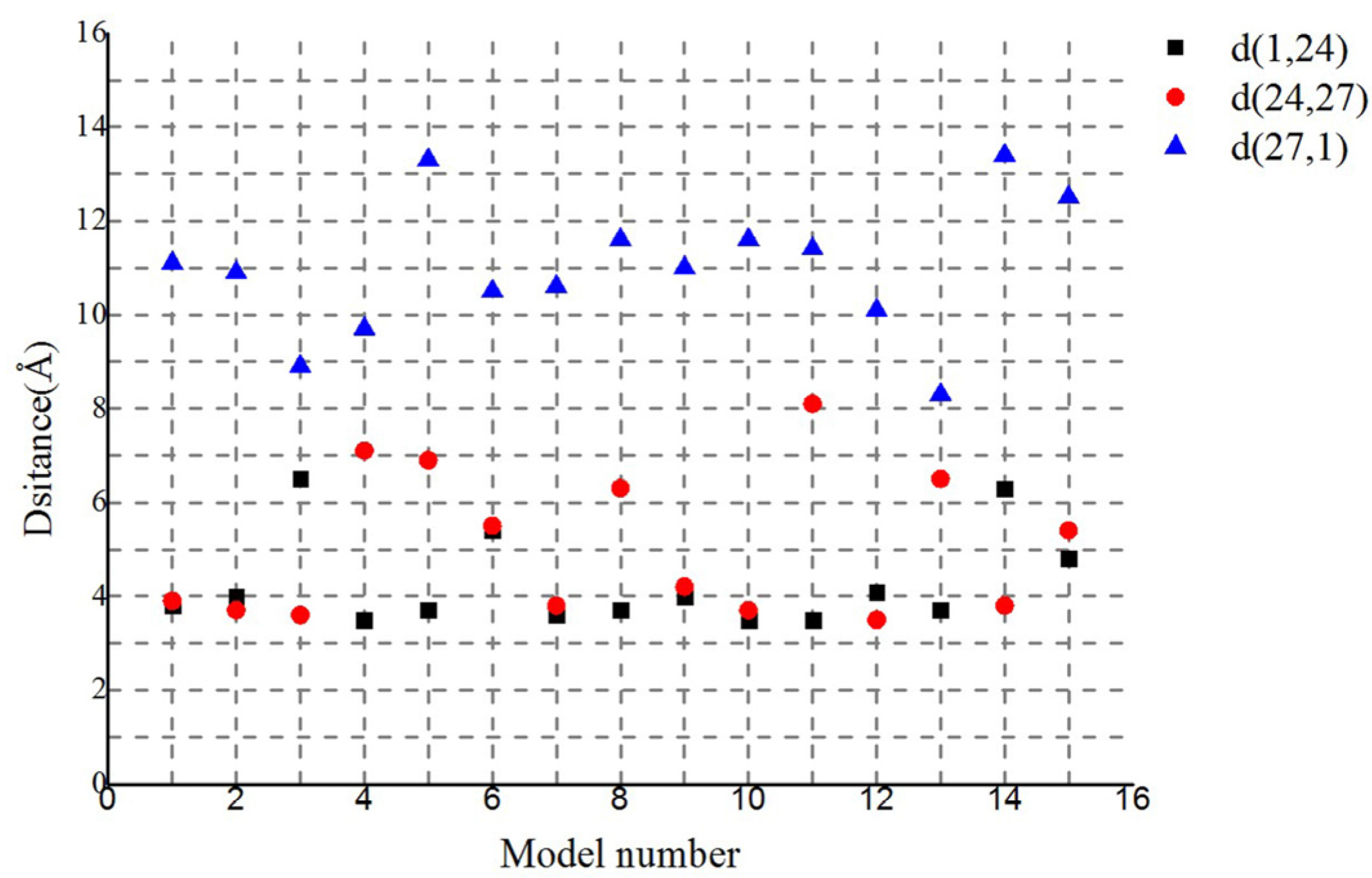
2.4. Co-Localization of Aromatic and Charged Residues in the G. uraniireducens Pilus
3. Materials and Methods
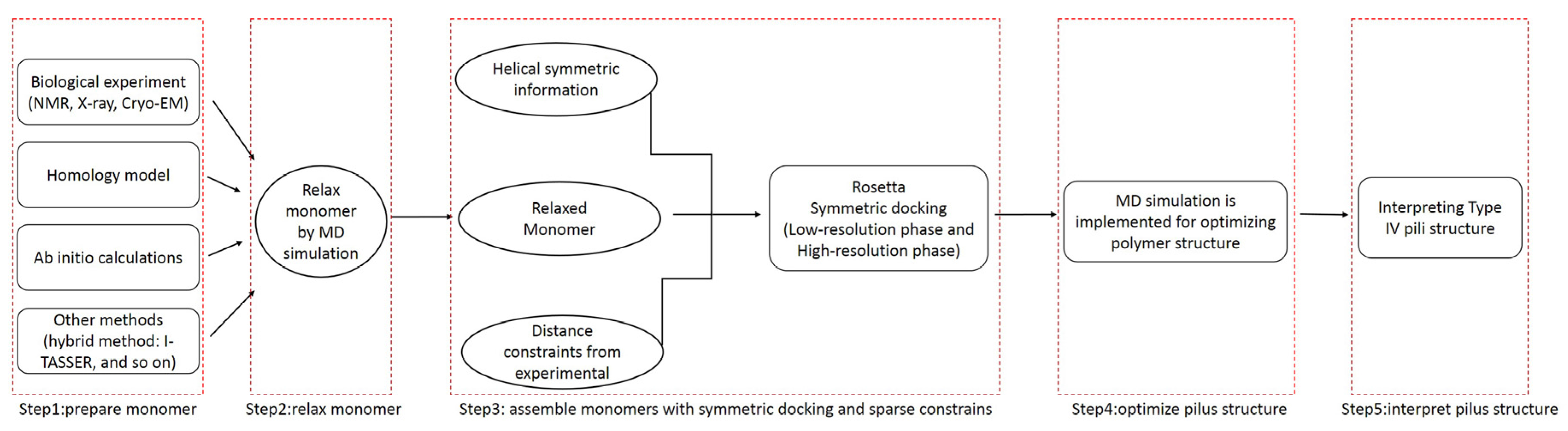
3.1. Overall Workflow
3.2. Molecular Dynamics Simulation
3.3. Symmetric Docking with Sparse Constrains
3.4. Energy Penalty Functions
3.5. Electrostatic Properties
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Malvankar, N.S.; Vargas, M.; Nevin, K.; Tremblay, P.L.; Evans-Lutterodt, K.; Nykypanchuk, D.; Martz, E.; Tuominen, M.T.; Lovley, D.R. Structural Basis for Metallic-Like Conductivity in Microbial Nanowires. mBio 2015, 6, e00084-15. [Google Scholar] [CrossRef] [PubMed]
- Reguera, G.; McCarthy, K.D.; Mehta, T.; Nicoll, J.S.; Tuominen, M.T.; Lovley, D.R. Extracellular electron transfer via microbial nanowires. Nature 2005, 435, 1098–1101. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Adhikari, R.Y.; Malvankar, N.S.; Ward, J.E.; Woodard, T.L.; Nevin, K.P.; Lovley, D.R. Expressing the Geobacter metallireducens PilA in Geobacter sulfurreducens Yields Pili with Exceptional Conductivity. mBio 2017, 8, e02203-16. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Adhikari, R.Y.; Malvankar, N.S.; Pi, S.; Ward, J.E.; Woodard, T.L.; Nevin, K.P.; Xia, Q.; Tuominen, M.T.; Lovley, D.R. Synthetic Biological Protein Nanowires with High Conductivity. Small 2016, 12, 4481–4485. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Adhikari, R.Y.; Malvankar, N.S.; Ward, J.E.; Nevin, K.P.; Woodard, T.L.; Smith, J.A.; Snoeyenbos-West, O.L.; Franks, A.E.; Tuominen, M.T.; et al. The Low Conductivity of Geobacter uraniireducens Pili Suggests a Diversity of Extracellular Electron Transfer Mechanisms in the Genus Geobacter. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.; Cisneros, D.A.; Nivaskumar, M.; Francetic, O. The type II secretion system—A dynamic fiber assembly nanomachine. Res. Microbiol. 2013, 164, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, A.; Milgotina, E.; Scanlon, K.; Craig, L.; Tainer, J.A.; Donnenberg, M.S. Structure of an essential type IV pilus biogenesis protein provides insights into pilus and type II secretion systems. J. Mol. Biol. 2012, 419, 110–124. [Google Scholar] [CrossRef] [PubMed]
- Burrows, L.L. A new twist in the assembly of type IV pilus-like fibers. Structure 2014, 22, 659–661. [Google Scholar] [CrossRef] [PubMed]
- Peabody, C.R.; Chung, Y.J.; Yen, M.R.; Vidal-Ingigliardi, D.; Pugsley, A.P.; Saier, M.J. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 2003, 149, 3051–3072. [Google Scholar] [CrossRef] [PubMed]
- Nivaskumar, M.; Francetic, O. Type II secretion system: A magic beanstalk or a protein escalator. Biochim. Biophys. Acta 2014, 1843, 1568–1577. [Google Scholar] [CrossRef] [PubMed]
- Craig, L.; Volkmann, N.; Arvai, A.S.; Pique, M.E.; Yeager, M.; Egelman, E.H.; Tainer, J.A. Type IV pilus structure by cryo-electron microscopy and crystallography: Implications for pilus assembly and functions. Mol. Cell. 2006, 23, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Strom, M.S.; Lory, S. Amino acid substitutions in pilin of Pseudomonas aeruginosa. Effect on leader peptide cleavage, amino-terminal methylation, and pilus assembly. J. Biol. Chem. 1991, 266, 1656–1664. [Google Scholar] [PubMed]
- Chiang, P.; Habash, M.; Burrows, L.L. Disparate subcellular localization patterns of Pseudomonas aeruginosa Type IV pilus ATPases involved in twitching motility. J. Bacteriol. 2005, 187, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Chiang, P.; Sampaleanu, L.M.; Ayers, M.; Pahuta, M.; Howell, P.L.; Burrows, L.L. Functional role of conserved residues in the characteristic secretion NTPase motifs of the Pseudomonas aeruginosa type IV pilus motor proteins PilB, PilT and PilU. Microbiology 2008, 154, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Nivaskumar, M.; Bouvier, G.; Campos, M.; Nadeau, N.; Yu, X.; Egelman, E.H.; Nilges, M.; Francetic, O. Distinct docking and stabilization steps of the Pseudopilus conformational transition path suggest rotational assembly of type IV pilus-like fibers. Structure 2014, 22, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Nunn, D. Bacterial type II protein export and pilus biogenesis: More than just homologies. Trends Cell. Biol. 1999, 9, 402–408. [Google Scholar] [CrossRef]
- Cehovin, A.; Simpson, P.J.; McDowell, M.A.; Brown, D.R.; Noschese, R.; Pallett, M.; Brady, J.; Baldwin, G.S.; Lea, S.M.; Matthews, S.J.; et al. Specific DNA recognition mediated by a type IV pilin. Proc. Natl. Acad. Sci. USA 2013, 110, 3065–3070. [Google Scholar] [CrossRef] [PubMed]
- Methe, B.A.; Nelson, K.E.; Eisen, J.A.; Paulsen, I.T.; Nelson, W.; Heidelberg, J.F.; Wu, D.; Wu, M.; Ward, N.; Beanan, M.J.; et al. Genome of Geobacter sulfurreducens: Metal reduction in subsurface environments. Science 2003, 302, 1967–1969. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.E.; Young, N.D.; Aklujkar, M.; Lovley, D.R. Comparative genomic analysis of Geobacter sulfurreducens KN400, a strain with enhanced capacity for extracellular electron transfer and electricity production. Bmc Genom. 2012, 13, 471. [Google Scholar] [CrossRef] [PubMed]
- Craig, L.; Pique, M.E.; Tainer, J.A. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2004, 2, 363–378. [Google Scholar] [CrossRef] [PubMed]
- Helaine, S.; Carbonnelle, E.; Prouvensier, L.; Beretti, J.L.; Nassif, X.; Pelicic, V. PilX, a pilus-associated protein essential for bacterial aggregation, is a key to pilus-facilitated attachment of Neisseria meningitidis to human cells. Mol. Microbiol. 2005, 55, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Lovely, D.R. Electromicrobiology. Annu. Rev. Microbiol. 2012, 66, 391–409. [Google Scholar] [CrossRef] [PubMed]
- Milne, J.L.S.; Borgnia, M.J.; Bartesaghi, A.; Tran, E.E.H.; Earl, L.A.; Schauder, D.M.; Lengyel, J.; Pierson, J.; Patwardhan, A.; Subramaniam, S. Cryo-electron microscopy—A primer for the non-microscopist. FEBS J. 2013, 280, 28–45. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E. The revolution will not be crystallized: A new method sweeps through structural biology. Nature 2015, 525, 172–174. [Google Scholar] [CrossRef] [PubMed]
- Kolappan, S.; Coureuil, M.; Yu, X.; Nassif, X.; Egelman, E.H.; Craig, L. Structure of the Neisseria meningitidis Type IV pilus. Nat. Commun. 2016, 7, 13015. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Chuang, C.; Zhugayevych, A.; Tretiak, S.; Dahlquist, F.W.; Bazan, G.C. Inter-aromatic distances in Geobacter sulfurreducens pili relevant to biofilm charge transport. Adv. Mater. 2015, 27, 1908–1911. [Google Scholar] [CrossRef] [PubMed]
- Xiao, K.; Shu, C.; Yan, Q.; Sun, X. Predicting Homogeneous Pilus Structure from Monomeric Data and Sparse Constraints. BioMed Res. Int. 2015, 2015, 817134. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Baker, D. Macromolecular modeling with rosetta. Annu. Rev. Biochem. 2008, 77, 363–382. [Google Scholar] [CrossRef] [PubMed]
- Presutti, E. Scaling Limits in Statistical Mechanics and Microstructures in Continuum Mechanics; Springer: Berlin/Heidelberg, Germany, 2009; p. 467. [Google Scholar]
- Zhao, X.M.; Xia, L.Q.; Yang, X.P.; Peng, X.Y. Flexibility Analysis of Bacillus thuringiensis Cry1Aa. Biomed. Environ. Sci. 2015, 28, 634–641. [Google Scholar] [PubMed]
- Hospital, A.; Goni, J.R.; Orozco, M.; Gelpi, J.L. Molecular dynamics simulations: Advances and applications. Adv. Appl. Bioinform. Chem. 2015, 10, 37–47. [Google Scholar]
- Feliciano, G.T.; Steidl, R.J.; Reguera, G. Structural and functional insights into the conductive pili of Geobacter sulfurreducens revealed in molecular dynamics simulations. Phys. Chem. Chem. Phys. 2015, 17, 22217–22226. [Google Scholar] [CrossRef] [PubMed]
- Vargas, M.; Malvankar, N.S.; Tremblay, P.L.; Leang, C.; Smith, J.A.; Patel, P.; Snoeyenbos-West, O.; Nevin, K.P.; Lovley, D.R. Aromatic Amino Acids Required for Pili Conductivity and Long-Range Extracellular Electron Transport in Geobacter sulfurreducens. mBio 2013, 4, e105–e113. [Google Scholar] [CrossRef]
- Wormald, M.R.; Petrescu, A.J.; Pao, Y.L.; Glithero, A.; Elliott, T.; Dwek, R.A. Conformational studies of oligosaccharides and glycopeptides: Complementarity of NMR, X-ray crystallography, and molecular modelling. Chem. Rev. 2002, 102, 371–386. [Google Scholar] [CrossRef] [PubMed]
- Nurisso, A.; Daina, A.; Walker, R.C. A practical introduction to molecular dynamics simulations: Applications to homology modeling. In Homology Modeling. Methods in Molecular Biology (Methods and Protocols); Orry, A., Abagyan, R., Eds.; Humana Press: New York, NY, USA, 2011; Volume 857, pp. 137–173. [Google Scholar] [PubMed]
- Andre, I.; Bradley, P.; Wang, C.; Baker, D. Prediction of the structure of symmetrical protein assemblies. Proc. Natl. Acad. Sci. USA 2007, 104, 17656–17661. [Google Scholar] [CrossRef] [PubMed]
- Shelobolina, E.S.; Vrionis, H.A.; Findlay, R.H.; Lovley, D.R. Geobacter uraniireducens sp. nov., isolated from subsurface sediment undergoing uranium bioremediation. Int. J. Syst. Evol. Microbiol. 2008, 58, 1075–1078. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Shu, C.; Xiao, K.; Yan, Q.; Sun, X. Comparative analysis of Type IV pilin in Desulfuromonadales. Front. Microbiol. 2016, 7, 2080. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.; Francetic, O.; Nilges, M. Modeling pilus structures from sparse data. J. Struct. Biol. 2011, 173, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.; Nilges, M.; Cisneros, D.A.; Francetic, O. Detailed structural and assembly model of the type II secretion pilus from sparse data. Proc. Natl. Acad. Sci. USA 2010, 107, 13081–13086. [Google Scholar] [CrossRef] [PubMed]
- Aas, F.E.; Wolfgang, M.; Frye, S.; Dunham, S.; Lovold, C.; Koomey, M. Competence for natural transformation in Neisseria gonorrhoeae: Components of DNA binding and uptake linked to type IV pilus expression. Mol. Microbiol. 2002, 46, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Shih, C.; Museth, A.K.; Abrahamsson, M.; Blanco-Rodriguez, A.M.; Di Bilio, A.J.; Sudhamsu, J.; Crane, B.R.; Ronayne, K.L.; Towrie, M.; Vlcek, A., Jr.; et al. Tryptophan-accelerated electron flow through proteins. Science 2008, 320, 1760–1762. [Google Scholar] [CrossRef] [PubMed]
- Page, C.C.; Moser, C.C.; Chen, X.; Dutton, P.L. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature 1999, 402, 47–52. [Google Scholar] [PubMed]
- DeLano, W.L. PyMOL: An open-source molecular graphics tool. CCP4 Newsletter On Protein Crystallography 2002, 40, 82–92. [Google Scholar]
- Dijkstra, E.W. A note on two problems in connexion with graphs. Numer. Math. 1959, 1, 269–271. [Google Scholar] [CrossRef]
- Xiao, K.; Malvankar, N.S.; Shu, C.; Martz, E.; Lovley, D.R.; Sun, X. Low Energy Atomic Models Suggesting a Pilus Structure that could Account for Electrical Conductivity of Geobacter sulfurreducens Pili. Sci. Rep.-UK 2016, 6, 23385. [Google Scholar] [CrossRef] [PubMed]
- Voityuk, A.A. Long-Range Electron Transfer in Biomolecules. Tunneling or Hopping? J. Phys. Chem. B 2011, 115, 12202–12207. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Darden, T.; Cheatham, T.; Simmerling, C.; Wang, J.; Duke, R.E.; Luo, R.; Walker, R.C.; Zhang, W.; Merz, K.; et al. Amber 12; University of California: San Francisco, CA, USA, 2012. [Google Scholar]
- Perez, A.; MacCallum, J.L.; Brini, E.; Simmerling, C.; Dill, K.A. Grid-Based Backbone Correction to the ff12SB Protein Force Field for Implicit-Solvent Simulations. J. Chem. Theory Comput. 2015, 11, 4770–4779. [Google Scholar] [CrossRef] [PubMed]
- Roe, D.R.; Cheatham, T.R. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Giltner, C.L.; Nguyen, Y.; Burrows, L.L. Type IV pilin proteins: Versatile molecular modules. Microbiol. Mol. Biol. Rev. 2012, 76, 740–772. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Lv, Q.; Yang, L.; Qian, P. Application of Affinity Propagation Clustering Algorithm in Protein Structure Prediction. Microelectr. Comput. 2010, 27, 154–157. [Google Scholar]
- Donald, J.E.; Kulp, D.W.; DeGrado, W.F. Salt bridges: Geometrically specific, designable interactions. Proteins 2011, 79, 898–915. [Google Scholar] [CrossRef] [PubMed]
- Unni, S.; Huang, Y.; Hanson, R.M.; Tobias, M.; Krishnan, S.; Li, W.W.; Nielsen, J.E.; Baker, N.A. Web servers and services for electrostatics calculations with APBS and PDB2PQR. J. Comput. Chem. 2011, 32, 1488–1491. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Tang, J.; Zou, Q. Local-DPP: An improved DNA-binding protein prediction method by exploring local evolutionary information. Inf. Sci. 2017, 384, 135–144. [Google Scholar] [CrossRef]
- Su, R.; Zhang, C.; Pham, T.D.; Davey, R.; Bischof, L.; Vallotton, P.; Lovell, D.; Hope, S.; Schmoelzl, S.; Sun, C. Detection of tubule boundaries based on circular shortest path and polar-transformation of arbitrary shapes. J. Microsc. 2016, 264, 127–142. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the structures GC1–4 and NM1–4 are available from the authors. |


| GC1 | GC2 | GC3 | GC4 | |
|---|---|---|---|---|
| GC1 | 0 | 2.739 | 3.290 | 2.308 |
| GC2 | - | 0 | 2.386 | 0.672 |
| GC3 | - | - | 0 | 2.343 |
| GC4 | - | - | - | 0 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shu, C.; Xiao, K.; Cao, C.; Ding, D.; Sun, X. Predicting and Interpreting the Structure of Type IV Pilus of Electricigens by Molecular Dynamics Simulations. Molecules 2017, 22, 1342. https://doi.org/10.3390/molecules22081342
Shu C, Xiao K, Cao C, Ding D, Sun X. Predicting and Interpreting the Structure of Type IV Pilus of Electricigens by Molecular Dynamics Simulations. Molecules. 2017; 22(8):1342. https://doi.org/10.3390/molecules22081342
Chicago/Turabian StyleShu, Chuanjun, Ke Xiao, Changchang Cao, Dewu Ding, and Xiao Sun. 2017. "Predicting and Interpreting the Structure of Type IV Pilus of Electricigens by Molecular Dynamics Simulations" Molecules 22, no. 8: 1342. https://doi.org/10.3390/molecules22081342





