Enzymatic Synthesis of Unnatural Ginsenosides Using a Promiscuous UDP-Glucosyltransferase from Bacillus subtilis
Abstract
:1. Introduction
2. Results
2.1. Heterologous Expression and Purification of UGT109A1
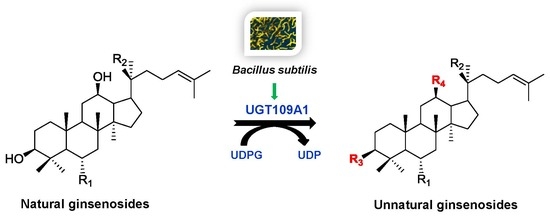
2.2. In Vitro Glycosylation of Ginsenosides by UGT109A1
2.3. Structure Identification of the Glycosylated Products
2.4. Optimization of Reaction Conditions for UGT109A1
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Gene Cloning, Expression and Purification of UGT109A1
4.3. In Vitro Activity Assay of UGT109A1
4.4. Effects of Temperature, pH and Metal Ions on the Activity of UGT109A1
4.5. HPLC Analysis of the Glycosylated Products
4.6. Scale-Up Reactions and Accumulation of the Glycosylated Products
4.7. HPLC-ESI-MS and NMR Analysis of the Glycosylated Products
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hemmerly, T.E. A ginseng farm in Lawrence County, Tennessee. Econ. Bot. 1977, 31, 160–162. [Google Scholar] [CrossRef]
- Shibata, S. Chemistry and tumor preventing activities of ginseng saponins and some related triterpenoid compounds. J. Korean Med. Sci. 2001, 16, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Yun, T.K. Panax ginseng—A non-organ-specific tumor preventive? Lancet Oncol. 2001, 2, 49–55. [Google Scholar] [CrossRef]
- Ahn, S.; Siddiqi, M.H.; Noh, H.Y.; Kim, Y.J.; Kim, Y.J.; Jin, C.G.; Yang, D.C. Anti-inflammatory activity of ginsenosides in LPS-stimulated RAW 264.7 cells. Sci. Bull. 2015, 60, 773–784. [Google Scholar] [CrossRef]
- Mathiyalagan, R.; Subramaniyam, S.; Kim, Y.J.; Kim, Y.C.; Yang, D.C. Ginsenoside compound K-bearing glycol chitosan conjugates: Synthesis, physicochemical characterization, and in vitro biological studies. Carbohydr. Polym. 2014, 112, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.X.; Yu, N.J.; Yang, X.H. The study of ginsenoside on PPARγ expression of mononuclear macrophage in type 2 diabetes. Mol. Biol. Rep. 2010, 37, 2975–2979. [Google Scholar] [CrossRef] [PubMed]
- Shin, B.K.; Kwon, S.W.; Park, J.H. Chemical diversity of ginseng saponins from Panax ginseng. J. Ginseng Res. 2015, 16, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Park, M.T.; Cha, H.J.; Jeong, J.W.; Lee, H.Y.; Kim, S.I.; Baek, N.I.; Kim, O.K.; Kim, K.W. Anti-invasive activity of ginsenoside Rh1 and Rh2 in the HT1080 cells. J. Ginseng Res. 1998, 22, 216–221. [Google Scholar]
- Lee, Y.J.; Jin, Y.R.; Lim, W.C.; Ji, S.M.; Choi, S.; Jang, S.; Lee, S. A ginsenoside-Rh1, a component of ginseng saponin, activates estrogen receptor in human breast carcinoma MCF-7 cells. J. Steroid Biochem. Mol. Biol. 2003, 84, 463–468. [Google Scholar] [CrossRef]
- Jung, J.S.; Shin, J.A.; Park, E.M.; Lee, J.E.; Kang, Y.S.; Min, S.W.; Kim, D.H.; Hyun, J.W.; Shin, C.Y.; Kim, H.S. Anti-inflammatory mechanism of ginsenoside Rh1 in lipopolysaccharide-stimulated microglia: Critical role of the protein kinase A pathway and hemeoxygenase-1 expression. J. Neurochem. 2010, 115, 1668–1680. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Fan, Y.; Wei, W.; Wang, P.P.; Liu, Q.F.; Wei, Y.J.; Zhang, L.; Zhao, G.P.; Yue, J.M.; Zhou, Z.H. Production of bioactive ginsenoside compound K in metabolically engineered yeast. Cell Res. 2014, 24, 770–773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.P.; Wei, Y.J.; Fan, Y.; Liu, Q.F.; Wei, W.; Yang, C.S.; Zhang, L.; Zhao, G.P.; Yue, J.M.; Yan, Y.; et al. Production of bioactive ginsenosides Rh2 and Rg3 by metabolically engineered yeasts. Metab. Eng. 2015, 29, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Wang, P.P.; Wei, Y.J.; Liu, Q.F.; Yang, C.F.; Zhao, G.P.; Yue, J.M.; Yan, Y.; Zhou, Z.H. Characterization of Panax ginseng UDP-glycosyltransferases catalyzing protopanaxatriol and biosyntheses of bioactive ginsenosides F1 and Rh1 in metabolically engineered yeasts. Mol. Plant. 2015, 8, 1412–1424. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.G.; Yang, S.M.; Kim, S.Y.; Cha, M.N.; Ahn, J.H. Biosynthesis and production of glycosylated flavonoids in Escherichia coli: Current state and perspectives. Appl. Microbiol. Biotechnol. 2015, 99, 2979–2988. [Google Scholar] [CrossRef] [PubMed]
- Simkhada, D.; Kurumbang, N.P.; Lee, H.C.; Sohng, J.K. Exploration of glycosylated flavonoids from metabolically engineered E. coli. Biotechnol. Bioprocess. Eng. 2010, 15, 754–760. [Google Scholar] [CrossRef]
- Pandey, R.P.; Gurung, R.B.; Parajuli, P.; Koirala, N.; Tuoi, L.T.; Sohng, J.K. Assessing acceptor substrate promiscuity of YjiC-mediated glycosylation toward flavonoids. Carbohydr. Res. 2014, 393, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.L.; Dang, L.Z.; Zhang, K.Q.; Liang, L.M.; Li, G.H. Cloning and heterologous expression of UDP-glycosyltransferase genes from Bacillus subtilis and its application in the glycosylation of ginsenoside Rh1. Lett. Appl. Microbiol. 2015, 60, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Gurung, R.B.; Kim, E.H.; Oh, T.J.; Sohng, J.K. Enzymatic synthesis of apigenin glucosides by glucosyltransferase (YjiC) from Bacillus licheniformis DSM 13. Mol. Cells 2013, 36, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.H.; Li, J.; Yao, P.Y.; Zhu, Y.M.; Men, Y.; Zeng, Y.; Yang, J.G.; Sun, Y.X. Exploiting the aglycon promiscuity of glycosyltransferase Bs-YjiC from Bacillus subtilis and its application in synthesis of glycosides. J. Biotechnol. 2017, 248, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.C.; Hu, Z.F.; Zhang, T.T.; Gong, T.; Chen, J.J.; Zhu, P.; Li, Y.; Yang, J.L. Production of a bioactive unnatural ginsenoside by metabolically engineered yeasts based on a new UDP-glycosyltransferase from Bacillus subtilis. Metab. Eng. 2017, 44, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.H.; Liu, C.; Li, J.; Dong, C.X.; Yang, J.G.; Dai, Z.B.; Zhang, X.L.; Sun, Y.X. One-pot synthesis of ginsenoside Rh2 and bioactive unnatural ginsenoside by coupling promiscuous glycosyltransferase from Bacillus subtilis 168 to sucrose synthase. J. Agric. Food Chem. 2018, 66, 2830–2837. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.H.; Li, J.; Yang, J.G.; Zhu, Y.M.; Men, Y.; Zeng, Y.; Cai, Y.; Dong, C.X.; Dai, Z.B.; Zhang, X.L.; et al. Use of a promiscuous glycosyltransferase from Bacillus subtilis 168 for the enzymatic synthesis of novel protopanaxatriol-type ginsenosides. J. Agric. Food Chem. 2018, 66, 943–949. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.T.; Shao, Z.H.; Hoek, T.L.V.; Chang, W.T.; Li, J.; Mehendale, S.; Wang, C.Z.; Hsu, C.W.; Becker, L.B.; Yin, J.J.; et al. Antioxidant effects of ginsenoside Re in cardiomyocytes. Eur. J. Pharmacol. 2006, 532, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Dang, D.K.; Shin, E.J.; Kim, D.J.; Tran, H.Q.; Jeong, J.H.; Jang, C.G.; Nah, S.Y.; Jeong, J.H.; Byun, J.K.; Ko, S.K.; et al. Ginsenoside Re protects methamphetamine-induced dopaminergic neurotoxicity in mice via upregulation of dynorphin-mediated κ-opioid receptor and downregulation of substance P-mediated neurokinin 1 receptor. J. Neuroinflammation. 2018, 15, 52. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.; Siddiqi, M.H.; Aceituno, V.C.; Simu, S.Y.; Yang, D.C. Suppression of MAPKs/NF-kB activation induces intestinal anti-inflammatory action of ginsenoside Rf in HT-29 and RAW264.7 cells. Immunol. Invest. 2016, 45, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Tam, D.N.H.; Truong, D.H.; Nguyen, T.T.H.; Quynh, L.N.; Tran, L.; Nguyen, H.D.; Shamandy, B.E.; Le, T.M.H.; Tran, D.K.; Sayed, D.; et al. Ginsenoside Rh1: A systematic review of its pharmacological properties. Planta Med. 2018, 84, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Zhang, M.; Feng, L.; Zhou, Y.; Li, L.; Ban, L. Protective effects of notoginsenoside R1 on cerebral ischemia-reperfusion injury in rats. Exp. Ther. Med. 2017, 14, 6012–6016. [Google Scholar] [CrossRef] [PubMed]
- Harle, J.; Bechthold, A. The power of glycosyltransferases to generate bioactive natural compounds. In Methods in Enzymology; Hopwood, D.A., Ed.; Academic Press: New York, NY, USA, 2009; Volume 458, pp. 309–333. [Google Scholar]
- Pandey, R.P.; Parajuli, P.; Shin, J.Y.; Lee, J.; Lee, S.; Hong, Y.S.; Park, Y.; Kim, J.S.; Sohnga, J.K. Enzymatic biosynthesis of novel resveratrol glucoside and glycoside derivatives. Appl. Environ. Microbiol. 2014, 80, 7235–7243. [Google Scholar] [CrossRef] [PubMed]
- Parajuli, P.; Pandey, R.P.; Koirala, N.; Yoon, Y.J.; Kim, B.G.; Sohng, J.K. Enzymatic synthesis of epothilone A glycosides. AMB Express 2014, 4, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Fan, S.; Chen, R.; Xie, K.; Yin, S.; Sun, L.; Liu, J.; Yang, L.; Kong, J.; Yang, Z.; et al. Probing and engineering key residues for bis-C-glycosylation and promiscuity of a C-glycosyltransferase. ACS Catal. 2018, 8, 4917–4927. [Google Scholar] [CrossRef]
- Wang, D.D.; Jin, Y.; Wang, C.; Kim, Y.J.; Perez, Z.E.J.; Baek, N.I.; Mathiyalagan, R.; Markus, J.; Yang, D.C. Rare ginsenoside Ia synthesized from F1 by cloning and overexpression of the UDP-glycosyltransferase gene from Bacillus subtilis: Synthesis, characterization, and in vitro melanogenesis inhibition activity in BL6B16 cells. J. Ginseng Res. 2018, 42, 42–49. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |







© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.-T.; Gong, T.; Hu, Z.-F.; Gu, A.-D.; Yang, J.-L.; Zhu, P. Enzymatic Synthesis of Unnatural Ginsenosides Using a Promiscuous UDP-Glucosyltransferase from Bacillus subtilis. Molecules 2018, 23, 2797. https://doi.org/10.3390/molecules23112797
Zhang T-T, Gong T, Hu Z-F, Gu A-D, Yang J-L, Zhu P. Enzymatic Synthesis of Unnatural Ginsenosides Using a Promiscuous UDP-Glucosyltransferase from Bacillus subtilis. Molecules. 2018; 23(11):2797. https://doi.org/10.3390/molecules23112797
Chicago/Turabian StyleZhang, Ting-Ting, Ting Gong, Zong-Feng Hu, An-Di Gu, Jin-Ling Yang, and Ping Zhu. 2018. "Enzymatic Synthesis of Unnatural Ginsenosides Using a Promiscuous UDP-Glucosyltransferase from Bacillus subtilis" Molecules 23, no. 11: 2797. https://doi.org/10.3390/molecules23112797