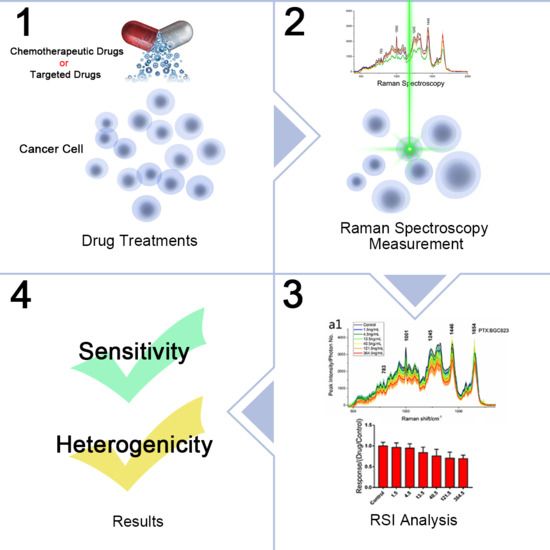
Anti-Cancer Drug Sensitivity Assay with Quantitative Heterogeneity Testing Using Single-Cell Raman Spectroscopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Confocal Raman Spectroscopy
2.2. Cell Culture
2.3. Anti-Cancer Drug Treatment of Cancer Cells
2.4. RS Measurements
2.5. CCK-8 (MTT) Assay and Direct Cell Counting
2.6. In Vitro Growth Inhibition Assays
2.7. Quantitative Measurements of the Heterogeneous Drug Responses
2.8. Experimental Consistency Control
2.9. Data Processing
3. Results and Discussion
3.1. Chemotherapy Drug Dose-Effect Assay
3.2. Quantification of Heterogeneous Drug Responses
3.3. Targeted Drug Growth/Inhibitor Evaluations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Samson, D.J.; Seidenfeld, J.; Ziegler, K.; Aronson, N. Chemotherapy sensitivity and resistance assays: A systematic review. J. Clin. Oncol. 2004, 22, 3618–3630. [Google Scholar] [CrossRef] [PubMed]
- Ulukaya, E. Drug response assay: An increasing trend in designation of tailored-chemotherapy for more rational management of cancer patients. Adv. Mol. Med. 2006, 2, 53–58. [Google Scholar]
- Lu, D.-Y.; Lu, T.-R.; Ding, J.; Xu, B.; Che, J.-Y.; Wu, H.-Y. Anticancer drug sensitivity testing, a historical review and future perspectives. Curr. Drug Ther. 2015, 10, 11. [Google Scholar] [CrossRef]
- Bedard, P.L.; Hansen, A.R.; Ratain, M.J.; Siu, L.L. Tumour heterogeneity in the clinic. Nature 2013, 501, 355–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meacham, C.E.; Morrison, S.J. Tumour heterogeneity and cancer cell plasticity. Nature 2013, 501, 328–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Junttila, M.R.; de Sauvage, F.J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 2013, 501, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Burrell, R.A.; McGranahan, N.; Bartek, J.; Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013, 501, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Kubota, T.; Egawa, T.; Otani, Y.; Furukawa, T.; Saikawa, Y.; Yoshida, M.; Watanabe, M.; Kumai, K.; Kitajima, M. Cancer chemotherapy chemosensitivity testing is useful in evaluating the appropriate adjuvant cancer chemotherapy for stages iii/iv gastric cancers without peritoneal dissemination. Anticancer Res. 2003, 23, 583–587. [Google Scholar] [PubMed]
- Lu, D.Y. Similarity of drug sensitivity test results on human pulmonary adenocarcinoma xenografts transplanted under the subrenal capsules between normal immunocompetent and immunodeficient athymic mice. Int. J. Pharm. Ther. 2010, 1, 106–109. [Google Scholar]
- Aamdal, S.; Fodstad, O.; Nesland, J.M.; Pihl, A. Characteristics of human tumour xenografts transplanted under the renal capsule of immunocompetent mice. Br. J. Cancer 1985, 51, 347–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alley, M.C.; Scudiero, D.A.; Monks, A.; Hursey, M.L.; Czerwinski, M.J.; Fine, D.L.; Abbott, B.J.; Mayo, J.G.; Shoemaker, R.H.; Boyd, M.R. Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res. 1988, 48, 589–601. [Google Scholar] [PubMed]
- Tominaga, H.; Ishiyama, M.; Ohseto, F.; Sasamoto, K.; Hamamoto, T.; Suzuki, K.; Watanabe, M. A water-soluble tetrazolium salt useful for colorimetric cell viability assay. Anal. Commun. 1999, 36, 47–50. [Google Scholar] [CrossRef]
- Xiao, Y.; Li, J.-D.; Shi, H.-L.; Liu, J.-H.; Feng, Y.-L.; Li, M.-D. Predictive value of in vitro mtt assay chemosensitivity test of cytotoxic drug activity in cervical cancer. Ai Zheng 2007, 26, 386–389. [Google Scholar] [PubMed]
- Debiec-Rychter, M.; Dumez, H.; Judson, I.; Wasag, B.; Verweij, J.; Brown, M.; Dimitrijevic, S.; Sciot, R.; Stul, M.; Vranck, H.; et al. Use of c-kit/pdgfra mutational analysis to predict the clinical response to imatinib in patients with advanced gastrointestinal stromal tumours entered on phase i and ii studies of the eortc soft tissue and bone sarcoma group. Eur. J. Cancer 2004, 40, 689–695. [Google Scholar] [CrossRef] [PubMed]
- Demonty, G.; Bernard-Marty, C.; Puglisi, F.; Mancini, I.; Piccart, M. Progress and new standards of care in the management of her-2 positive breast cancer. Eur. J. Cancer 2007, 43, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Hammoud, M.K.; Yosef, H.K.; Lechtonen, T.; Aljakouch, K.; Schuler, M.; Alsaidi, W.; Daho, I.; Maghnouj, A.; Hahn, S.; El-Mashtoly, S.F.; et al. Raman micro-spectroscopy monitors acquired resistance to targeted cancer therapy at the cellular level. Sci. Rep. 2018, 8, 15278. [Google Scholar] [CrossRef] [PubMed]
- Austin, L.A.; Osseiran, S.; Evans, C.L. Raman technologies in cancer diagnostics. Analyst 2016, 141, 476–503. [Google Scholar] [CrossRef] [PubMed]
- Schie, I.W.; Huser, T. Methods and applications of raman microspectroscopy to single-cell analysis. Appl. Spectrosc. 2013, 67, 813–828. [Google Scholar] [CrossRef] [PubMed]
- Visvader, J.E.; Lindeman, G.J. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat. Rev. Cancer 2008, 8, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Laterra, J. Cancer stem cells: Distinct entities or dynamically regulated phenotypes? Cancer Res. 2012, 72, 576–580. [Google Scholar] [CrossRef] [PubMed]
- Magee, J.A.; Piskounova, E.; Morrison, S.J. Cancer stem cells: Impact, heterogeneity, and uncertainty. Cancer Cell 2012, 21, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.W.; Lieu, D.K.; Huser, T.; Li, R.A. Label-free separation of human embryonic stem cells and their cardiac derivatives using raman spectroscopy. Anal. Chem. 2009, 81, 1324–1331. [Google Scholar] [CrossRef] [PubMed]
- Harkness, L.; Novikov, S.M.; Beermann, J.; Bozhevolnyi, S.I.; Kassem, M. Identification of abnormal stem cells using raman spectroscopy. Stem Cells Dev. 2012, 21, 2152–2159. [Google Scholar] [CrossRef] [PubMed]
- Manikandan, M.; Nasser Abdelhamid, H.; Talib, A.; Wu, H.F. Facile synthesis of gold nanohexagons on graphene templates in raman spectroscopy for biosensing cancer and cancer stem cells. Biosens. Bioelectron. 2014, 55, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Jung, G.B.; Lee, Y.J.; Lee, G.; Park, H.K. A simple and rapid detection of tissue adhesive-induced biochemical changes in cells and DNA using raman spectroscopy. Biomed. Opt. Express 2013, 4, 2673–2682. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Tao, Z.; Ai, M.; Peng, L.; Wang, G.; He, B.; Li, Y.-Q. Raman spectroscopic analysis of apoptosis of single human gastric cancer cells. Vib. Spectrosc. 2009, 50, 193–197. [Google Scholar] [CrossRef]
- Buckmaster, R.; Asphahani, F.; Thein, M.; Xu, J.; Zhang, M. Detection of drug-induced cellular changes using confocal raman spectroscopy on patterned single-cell biosensors. Analyst 2009, 134, 1440–1446. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Shi, H.; Feng, S.; Chen, W.; Yu, Y.; Lin, D.; Chen, R. Confocal raman spectroscopic analysis of the cytotoxic response to cisplatin in nasopharyngeal carcinoma cells. Anal. Methods 2013, 5, 260–266. [Google Scholar] [CrossRef]
- Aljakouch, K.; Lechtonen, T.; Yosef, H.K.; Hammoud, M.K.; Alsaidi, W.; Kotting, C.; Mugge, C.; Kourist, R.; El-Mashtoly, S.F.; Gerwert, K. Raman microspectroscopic evidence for the metabolism of a tyrosine kinase inhibitor, neratinib, in cancer cells. Angew. Chem. Int. Ed. 2018, 57, 7250–7254. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Parker, B.A.; Schwab, R.; Kurzrock, R. Her2 aberrations in cancer: Implications for therapy. Cancer Treat. Rev. 2014, 40, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Takegawa, N.; Nonagase, Y.; Yonesaka, K.; Sakai, K.; Maenishi, O.; Ogitani, Y.; Tamura, T.; Nishio, K.; Nakagawa, K.; Tsurutani, J. Ds-8201a, a new her2-targeting antibody-drug conjugate incorporating a novel DNA topoisomerase i inhibitor, overcomes her2-positive gastric cancer t-dm1 resistance. Int. J. Cancer 2017, 141, 1682–1689. [Google Scholar] [CrossRef] [PubMed]
- Bang, Y.-J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of her2-positive advanced gastric or gastro-oesophageal junction cancer (toga): A phase 3, open-label, randomised controlled trial. Lancet 2010, 376, 687–697. [Google Scholar] [CrossRef]
- Fujimoto-Ouchi, K.; Sekiguchi, F.; Yasuno, H.; Moriya, Y.; Mori, K.; Tanaka, Y. Antitumor activity of trastuzumab in combination with chemotherapy in human gastric cancer xenograft models. Cancer Chemother. Pharm. 2007, 59, 795–805. [Google Scholar] [CrossRef] [PubMed]
- Cejka, D.; Preusser, M.; Woehrer, A.; Sieghart, W.; Strommer, S.; Werzowa, J.; Fuereder, T.; Wacheck, V. Everolimus (rad001) and anti-angiogenic cyclophosphamide show long-term control of gastric cancer growth in vivo. Cancer Biol. Ther. 2014, 7, 1377–1385. [Google Scholar] [CrossRef]
- Crump, K.S.; Hoel, D.G.; Langley, C.H.; Peto, R. Fundamental carcinogenic processes and their implications for low dose risk assessment. Cancer Res. 1976, 36, 2973–2979. [Google Scholar] [PubMed]
- Di Veroli, G.Y.; Fornari, C.; Goldlust, I.; Mills, G.; Koh, S.B.; Bramhall, J.L.; Richards, F.M.; Jodrell, D.I. An automated fitting procedure and software for dose-response curves with multiphasic features. Sci. Rep. 2015, 5, 14701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, H.; Zhang, Y.; Ye, A. Single-cell discrimination based on optical tweezers raman spectroscopy. Chin. Sci. Bull. 2013, 58, 2594–2600. [Google Scholar] [CrossRef]
- Zhang, Y.; Jin, L.; Xu, J.; Yu, Y.; Shen, L.; Gao, J.; Ye, A. Dynamic characterization of drug resistance and heterogeneity of the gastric cancer cell bgc823 using single-cell raman spectroscopy. Analyst 2018, 143, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Tu, L.; Zhang, L. Comparison of paclitaxel-induced apoptosis in human gastric cancer bgc-823 and sgc-7901 cell. Anhui Med. Pharm. J. 2010, 14, 141–142. [Google Scholar]
- Wang, P.; Wang, C.; Wei, S.; Wang, C.; Hou, H. Establishment of cistplatin-resistant bgc 823/cddp cells. Chongqing Med. 2011, 40, 323–325. [Google Scholar]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. Past: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 10. [Google Scholar]
- Kroemer, G.; Galluzzi, L.; Vandenabeele, P.; Abrams, J.; Alnemri, E.S.; Baehrecke, E.H.; Blagosklonny, M.V.; El-Deiry, W.S.; Golstein, P.; Green, D.R.; et al. Classification of cell death: Recommendations of the nomenclature committee on cell death 2009. Cell Death Differ. 2009, 16, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Danial, N.N.; Korsmeyer, S.J. Cell death: Critical control points. Cell 2004, 116, 205. [Google Scholar] [CrossRef]
- Kroemer, G.; Galluzzi, L.; Brenner, C. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 2007, 87, 99–163. [Google Scholar] [CrossRef] [PubMed]
- Owen, C.A.; Selvakumaran, J.; Notingher, I.; Jell, G.; Hench, L.L.; Stevens, M.M. In vitro toxicology evaluation of pharmaceuticals using raman micro-spectroscopy. J. Cell. Biochem. 2006, 99, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Notingher, I.; Selvakumaran, J.; Hench, L.L. New detection system for toxic agents based on continuous spectroscopic monitoring of living cells. Biosens. Bioelectron. 2004, 20, 780–789. [Google Scholar] [CrossRef] [PubMed]
- De Gelder, J.; De Gussem, K.; Vandenabeele, P.; Moens, L. Reference database of raman spectra of biological molecules. J. Raman Spectrosc. 2007, 38, 1133–1147. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, X.; Liu, Y.; Zhang, S.; Liu, J.; Ma, Y.; Zhang, J. Antitumor effect of the mtor inhibitor everolimus in combination with trastuzumab on human breast cancer stem cells in vitro and in vivo. Tumor Biol. 2012, 33, 1349. [Google Scholar] [CrossRef] [PubMed]
- Jerusalem, G.; Fasolo, A.; Dieras, V.; Cardoso, F.; Bergh, J.; Vittori, L.; Zhang, Y.; Massacesi, C.; Sahmoud, T.; Gianni, L. Phase i trial of oral mtor inhibitor everolimus in combination with trastuzumab and vinorelbine in pre-treated patients with her2-overexpressing metastatic breast cancer. Breast Cancer Res. Treat. 2011, 125, 447–455. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |





| Test Groups | AUC | RSI-1001 cm−1 | CCK-8 (MTT) |
|---|---|---|---|
| PTX: BGC 823 | 0.9960 | 0.9995 | 0.9726 |
| DDP: SGC 7901 | 0.9721 | 0.9518 | 0.9961 |
| PTX: MGC 803 | 0.9424 | 0.8528 | 0.9761 |
| DDP + 5-Fu: AGS | 0.9531 | 0.9540 | 0.9949 |
| PTX + 5-Fu: MGC 803 | 0.9009 | 0.9009 | 0.8838 |
| Mean R2 | 0.9529 ± 0.0355 | 0.9315 ± 0.0565 | 0.9647 ± 0.0465 |
| Test Groups | AUC: MTT | RSI-1001 cm−1:MTT | AUC:RSI-1001 cm−1 |
|---|---|---|---|
| PTX: BGC823 | 0.9267 (p 1 = 0.008) | 0.9051 (p = 0.013) | 0.9956 (p < 0.000) |
| DDP: SGC7901 | 0.7352 (p = 0.096) | 0.7713 (p = 0.072) | 0.9961 (p < 0.000) |
| PTX: MGC803 | 0.9316 (p = 0.007) | 0.6963 (p = 0.124) | 0.9021 (p = 0.014) |
| DDP + 5Fu: AGS | 0.8832 (p = 0.019) | 0.8705 (p = 0.024) | 0.9986 (p < 0.000) |
| PTX + 5Fu: MGC803 | 0.8024 (p = 0.054) | 0.8171 (p = 0.047) | 0.9992 (p < 0.000) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Xu, J.; Yu, Y.; Shang, W.; Ye, A. Anti-Cancer Drug Sensitivity Assay with Quantitative Heterogeneity Testing Using Single-Cell Raman Spectroscopy. Molecules 2018, 23, 2903. https://doi.org/10.3390/molecules23112903
Zhang Y, Xu J, Yu Y, Shang W, Ye A. Anti-Cancer Drug Sensitivity Assay with Quantitative Heterogeneity Testing Using Single-Cell Raman Spectroscopy. Molecules. 2018; 23(11):2903. https://doi.org/10.3390/molecules23112903
Chicago/Turabian StyleZhang, Yong, Jingjing Xu, Yuezhou Yu, Wenhao Shang, and Anpei Ye. 2018. "Anti-Cancer Drug Sensitivity Assay with Quantitative Heterogeneity Testing Using Single-Cell Raman Spectroscopy" Molecules 23, no. 11: 2903. https://doi.org/10.3390/molecules23112903