Interactions of Bromocarbazoles with Human Serum Albumin Using Spectroscopic Methods
Abstract
:1. Introduction
2. Experimental Section
2.1. Materials
2.2. Fluorescence Spectrum
2.3. UV-Vis Absorbance Spectrum
2.4. Synchronous and 3D Fluorescence Spectra Measurements
2.5. CD Spectra
2.6. F.T-IR Spectrum Experiments
3. Results and Discussion
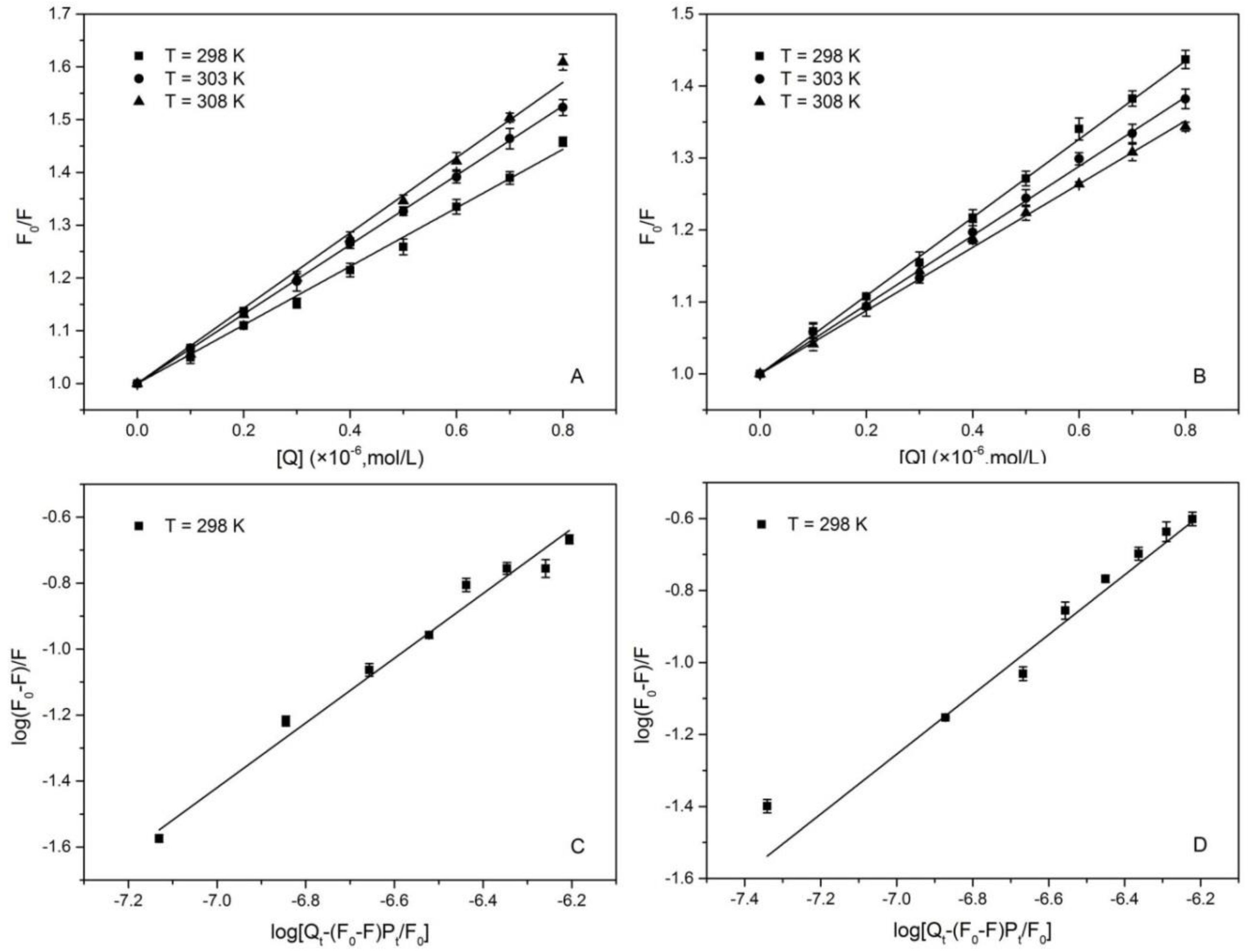
3.1. Fluorescence Quenching Mechanisms of 1,3,6,8-Tetrabromocarbazole and 3-Bromocarbazole Binding to HSA
3.2. Binding Constants and Location
3.3. Thermodynamic Parameters and Interaction Modes
3.4. Study of Conformational Changes of HSA
3.5. Fluorescence Resonance Energy Transfer
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Chen, W.; Xie, Z.; Wolschke, H.; Gandrass, J.; Koetke, D.; Winkelmann, M.; Ebinghaus, R. Quantitative determination of ultra-trace carbazoles in sediments in the coastal environment. Chemosphere 2016, 150, 586–595. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wang, W.; Liu, J.; Pan, D.; Tu, X.; Lv, P.; Wang, Y.; Cao, H.; Wang, Y.; Hua, R. Rapid biodegradation of the herbicide 2,4-dichlorophenoxyacetic acid by Cupriavidus gilardii T-1. J. Agric. Food Chem. 2017, 65, 3711–3720. [Google Scholar] [CrossRef] [PubMed]
- Parette, R.; McCrindle, R.; McMahon, K.S.; Pena-Abaurrea, M.; Reiner, E.; Chittim, B.; Riddell, N.; Voss, G.; Dorman, F.L.; Pearson, W.N. Halogenated indigo dyes: A likely source of 1,3,6,8-tetrabromocarbazole and some other halogenated carbazoles in the environment. Chemosphere 2015, 127, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.Y.; Hites, R.A. Identification of brominated carbazoles in sediment cores from Lake Michigan. Environ. Sci. Technol. 2005, 39, 9446–9451. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Chen, D.; Potter, D.; Rockne, K.J.; Sturchio, N.C.; Giesy, J.P.; Li, A. Polyhalogenated carbazoles in sediments of Lake Michigan: A New Discovery. Environ. Sci. Technol. 2014, 48, 12807–12815. [Google Scholar] [CrossRef] [PubMed]
- Beigoli, S.; Rad, A.S.; Askari, A.; Darban, R.A.; Chamani, J. Isothermal titration calorimetry and stopped flow circular dichroism investigations of the interaction between lomefloxacin and human serum albumin in the presence of amino acids. J. Biomol. Struct. Dyn. 2018, 1–53. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, L.; Zhu, M.; Xue, J.; Hua, R.; Li, Q.X. Comparative studies on biophysical interactions between gambogic acid and serum albumin via multispectroscopic approaches and molecular docking. J. Lumin. 2019, 205, 210–218. [Google Scholar] [CrossRef]
- Szkudlarek, A.; Pentak, D.; Ploch, A.; Pozycka, J.; Maciazek-Jurczyk, M. In Vitro Investigation of the interaction of tolbutamide and losartan with human serum albumin in hyperglycemia states. Molecules 2017, 22, 2249. [Google Scholar] [CrossRef] [PubMed]
- Abdelhameed, A.S.; Bakheit, A.H.; Almutairi, F.M.; AlRabiah, H.; Kadi, A.A. Biophysical and in silico studies of the interaction between the anti-viral agents acyclovir and penciclovir, and human serum albumin. Molecules 2017, 22, 1906. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Liu, W.; Li, Y.; Zhang, L.; Sun, Y. Determining the binding affinity and binding site of bensulfuron-methyl to human serum albumin by quenching of the intrinsic tryptophan fluorescence. J. Lumin. 2010, 130, 2013–2021. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Zhu, M.; Jiang, E.; Hua, R.; Li, Q.X. A simple and rapid turn on ESIPT fluorescent probe for colorimetric and ratiometric detection of biothiols in living cells. Sci. Rep. 2017, 7, 4377. [Google Scholar] [CrossRef] [PubMed]
- Rimac, H.; Dufour, C.; Debeljak, Ž.; Zorc, B.; Bojić, M. Warfarin and flavonoids do not share the same binding region in binding to the IIA subdomain of human serum albumin. Molecules 2017, 22, 1153. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, X.; Chen, X. Binding mechanism of Orange G to human serum albumin: Saturation transfer difference-NMR, spectroscopic and computational techniques. Dyes Pigm. 2013, 98, 212–220. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Jiang, E.; Zhu, M.; Wang, Z.; Fan, S.; Gao, Q.; Liu, S.; Li, Q.X.; Hua, R. A colorimetric and ratiometric dual-site fluorescent probe with 2,4-dinitrobenzenesulfonyl and aldehyde groups for imaging of aminothiols in living cells and zebrafish. Dyes Pigm. 2018, 156, 338–347. [Google Scholar] [CrossRef]
- Liang, G.W.; Chen, Y.C.; Wang, Y.; Wang, H.M.; Pan, X.Y.; Chen, P.H.; Niu, Q.X. Interaction between Saikosaponin D, Paeoniflorin, and Human Serum Albumin. Molecules 2018, 23, 249. [Google Scholar] [CrossRef] [PubMed]
- Na, R.; Zhu, M.; Fan, S.; Wang, Z.; Wu, X.; Tang, J.; Liu, J.; Wang, Y.; Hua, R. A simple and effective ratiometric fluorescent probe for the selective detection of cysteine and homocysteine in aqueous media. Molecules 2016, 21, 1023. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, M.; Jiang, L.; Song, L. New insight into molecular interaction of heavy metal pollutant-cadmium(II) with human serum albumin. Environ. Sci. Pollut. Res. 2014, 21, 6994–7005. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.Y.; Wang, Y.R.; Zhou, S.S.; Liu, Y. Investigation of the interaction between perfluorododecanoic acid and human serum albumin by multi-spectroscopic and molecular modeling techniques. Spectrosc. Spectral Anal. 2016, 36, 2330–2336. [Google Scholar]
- Zhu, M.; Wang, L.; Wang, Y.; Zhou, J.; Ding, J.; Li, W.; Xin, Y.; Fan, S.; Wang, Z.; Wang, Y. Biointeractions of herbicide atrazine with human serum albumin: UV-Vis, fluorescence and circular dichroism approaches. Int. J. Env. Res. Public Health 2018, 15, 116. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhu, M.; Liu, F.; Wu, X.; Pan, D.; Liu, J.; Fan, S.; Wang, Z.; Tang, J.; Na, R.; et al. Comparative studies of interactions between fluorodihydroquinazolin derivatives and human serum albumin with fluorescence spectroscopy. Molecules 2016, 21, 1373. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Diao, J.X.; Sun, Y.; Sun, Y. Bioevaluation of human serum albumin-hesperidin bioconjugate: Insight into protein vector function and conformation. J. Agric. Food Chem. 2012, 60, 7218–7228. [Google Scholar] [CrossRef] [PubMed]
- Peters, T., Jr. All About Albumin; Elsevier: San Diego, USA, 1995; pp. 319–413. [Google Scholar]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed; Springer: New York, NY, USA, 2006; pp. 456–457. [Google Scholar]
- Ding, X.; Suo, Z.; Sun, Q.; Gan, R.; Tang, P.; Hou, Q.; Wu, D.; Li, H. Study of the interaction of broad-spectrum antimicrobial drug sitafloxacin with human serum albumin using spectroscopic methods, molecular docking, and molecular dynamics simulation. J. Pharm. Biomed. Anal. 2018, 160, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, M.; Nusrat, S.; Alam, P.; Malik, S.; Chaturvedi, S.K.; Ajmal, M.R.; Abdelhameed, A.S.; Khan, R.H. Investigating the site selective binding of busulfan to human serum albumin: Biophysical and molecular docking approaches. Int. J. Biol. Macromol. 2018, 107, 1414–1421. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R.; Weber, G. Quenching of fluorescence by oxygen. A probe for structural fluctuations in macromolecules. Biochemistry 1973, 12, 4161–4170. [Google Scholar] [CrossRef] [PubMed]
- Soares, S.; Mateus, N.; Vd, F. Interaction of different polyphenols with bovine serum albumin (BSA) and human salivary alpha-amylase (HSA) by fluorescence quenching. J. Agric. Food Chem. 2007, 55, 6726–6735. [Google Scholar] [CrossRef] [PubMed]
- Bi, S.; Zhao, T.; Zhou, H.; Wang, Y.; Li, Z. Probing the interactions of bromchlorbuterol-HCl and phenylethanolamine A with HSA by multi-spectroscopic and molecular docking technique. J. Chem. Thermodyn. 2016, 97, 113–121. [Google Scholar] [CrossRef]
- Klotz, I.M. Ultraviolet absorption spectroscopy. Methods Mol. Biol. 1995, 40, 91–114. [Google Scholar] [CrossRef]
- Mandal, P.; Ganguly, T. Fluorescence spectroscopic characterization of the interaction of human adult hemoglobin and two isatins, 1-methylisatin and 1-phenylisatin: A comparative study. J. Phys. Chem. B 2009, 113, 14904–14913. [Google Scholar] [CrossRef] [PubMed]
- Sudlow, G.; Birkett, D.J.; Wade, D.N. The characterization of two specific drug binding sites on human serum albumin. Mol. Pharmacol. 1975, 11, 824–832. [Google Scholar] [PubMed]
- Sjoholm, I.; Ekman, B.; Kober, A.; Ljungstedt-Pahlman, I.; Seiving, B.; Sjodin, T. Binding of drugs to human serum albumin: XI. The specificity of three binding sites as studied with albumin immobilized in microparticles. Mol. Pharmacol. 1979, 16, 767–777. [Google Scholar] [PubMed]
- Ross, P.D.; Subramanian, S. Thermodynamics of protein association reactions—forces contributing to stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Li, Q.X. Pi-cation interactions in molecular recognition: Perspectives on pharmaceuticals and pesticides. J. Agric. Food Chem. 2018, 66, 3315–3323. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Wang, Y.; Liu, Y.; Xiang, Y. Exploration of interaction of canthaxanthin with human serum albumin by spectroscopic and molecular simulation methods. Luminescence 2018, 33, 425–432. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Yang, H.; Li, S.; Xu, K.; Wang, Q.; Huang, Y.; Li, H. Characterization of the interaction between acotiamide hydrochloride and human serum albumin: H-1 STD NMR spectroscopy, electrochemical measurement, and docking investigations. RSC Adv. 2016, 6, 61119–61128. [Google Scholar] [CrossRef]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Liu, D.; Zhang, Y.; Zhang, Z.; Li, H. Unravelling the binding mechanism of benproperine with human serum albumin: A docking, fluorometric, and thermodynamic approach. Eur. J. Med. Chem. 2018, 146, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.N.; Liu, J.Q.; He, W.Y.; Hu, Z.D.; Yao, X.J.; Chen, X.G. Probing the binding of scutellarin to human serum albumin by circular dichroism, fluorescence spectroscopy, FTIR, and molecular modeling method. Biomacromolecules 2004, 5, 1956–1961. [Google Scholar] [CrossRef] [PubMed]
- Bolattin, M.B.; Nandibewoor, S.T.; Joshi, S.D.; Dixit, S.R.; Chimatadar, S.A. Interaction of hydralazine with human serum albumin and effect of beta-Cyclodextrin on binding: Insights from spectroscopic and molecular docking techniques. Ind. Eng. Chem. Res. 2016, 55, 5454–5464. [Google Scholar] [CrossRef]
- Sinisi, V.; Forzato, C.; Cefarin, N.; Navarini, L.; Berti, F. Interaction of chlorogenic acids and quinides from coffee with human serum albumin. Food Chem. 2015, 168, 332–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, M.Q.; Wang, L.J.; Zhang, H.; Fan, S.S.; Wang, Z.; Li, Q.X.; Wang, Y.; Liu, S.Z. Interactions between tetrahydroisoindoline-1,3-dione derivatives and human serum albumin via multiple spectroscopy techniques. Environ. Sci. Pollut. Res. 2018, 25, 17735–17748. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Wang, Q.; Peng, X.; Li, H. The binding properties of metandienone and human serum albumin by comparing with other five similar compounds. J. Biochem. Mol. Toxicol. 2017, 31, 9. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Xu, H.; Zhu, Y.; Liu, J.; Wang, K.; Wang, P.; Shang, S.; Yi, X.; Wang, Z.; Shao, W.; et al. Understanding the fate of an anesthetic, nalorphine upon interaction with human serum albumin: A photophysical and mass-spectroscopy approach. RSC Adv. 2014, 4, 25410–25419. [Google Scholar] [CrossRef]
- Liu, B.M.; Zhang, J.; Hao, A.J.; Xu, L.; Wang, D.; Ji, H.; Sun, S.J.; Chen, B.-Q.; Liu, B. The increased binding affinity of curcumin with human serum albumin in the presence of rutin and baicalin: A potential for drug delivery system. Spectrochim. Acta. A Mol. Biomol. Spectrosc. 2016, 155, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, J.; Zhu, M.Q.; Wang, L.J.; Zen, X.; Fan, S.S.; Wang, Z.; Li, H.; Na, R.S.; Zhao, X.; et al. Biophysical characterization of interactions between falcarinol-type polyacetylenes and human serum albumin via multispectroscopy and molecular docking techniques. J. Lumin. 2018, 200, 111–119. [Google Scholar] [CrossRef]
- Cheung, H.C. Resonance Energy Transfer. Wiley: New York, NY, USA, 1999; pp. 259–279. [Google Scholar]
- Shaw, A.K.; Pal, S.K. Spectroscopic studies on the effect of temperature on pH-induced folded states of human serum albumin. J. Photochem. Photobiol. B Biol. 2008, 90, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Förster, T.; Sinanoglu, O. Modern Quantum Chemistry. Academic Press: New York, NY, USA, 1996. [Google Scholar]
- Yue, Y.; Sun, Y.; Dong, Q.; Liu, R.; Yan, X.; Zhang, Y.; Liu, J. Interaction of human serum albumin with novel imidazole derivatives studied by spectroscopy and molecular docking. Luminescence 2016, 31, 671–681. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |







| System | T (K) | KSV ± SD b (× 105·M−1) | Kq (× 1013·M−1·s−1) | R2 |
|---|---|---|---|---|
| 298 | 5.55 ± 0.02 | 5.55 | 0.998 | |
| HSA-3-bromocarbazole | 303 | 6.57 ± 0.01 | 6.57 | 0.999 |
| 308 | 7.13 ± 0.01 | 7.13 | 0.997 | |
| 298 | 5.43 ± 0.03 | 5.43 | 0.999 | |
| HSA-1,3,6,8-tetrabromocarbazole | 303 | 4.81 ± 0.04 | 4.81 | 0.998 |
| 308 | 4.40 ± 0.03 | 4.40 | 0.999 |
| Compound | T(K) | Ka ± SDb (×105·M−1) | n | R2 | ΔH (kJ·mol−1) | ΔG (kJ·mol−1) | ΔS (J·mol−1·K−1) |
|---|---|---|---|---|---|---|---|
| HSA-3-bromocarbazole | 298 | 2.18 ± 0.39 | 0.956 | 0.978 | −30.49 | ||
| 303 | 2.67 ± 0.15 | 0.847 | 0.988 | 24.65 | −31.41 | 185.02 | |
| 308 | 3.01 ± 0.20 | 0.78 | 0.984 | −32.33 | |||
| HAS-1,3,6,8-tetrabromocarbazole | 298 | 3.39 ± 0.40 | 0.703 | 0.948 | −31.57 | ||
| 303 | 3.05 ± 0.14 | 0.866 | 0.993 | −19.06 | −31.78 | 41.96 | |
| 308 | 2.64 ± 0.14 | 0.870 | 0.993 | −31.99 |
| System | Site Marker | Ka± SDb (×105 M−1) | R2 |
|---|---|---|---|
| HSA-3-bromocarbazole | Blank | 2.18 ± 0.39 | 0.978 |
| PB | 1.92 ± 0.15 | 0.993 | |
| FA | 1.15 ± 0.17 | 0.983 | |
| Dig | 1.62 ± 0.26 | 0.976 | |
| HSA-1,3,6,8-tetrabromocarbazole | Blank | 3.39 ± 0.40 | 0.948 |
| PB | 3.31 ± 0.19 | 0.989 | |
| FA | 2.78 ± 0.10 | 0.999 | |
| Dig | 3.12 ± 0.33 | 0.997 |
| Sample | Secondary Structure (%) | |||
|---|---|---|---|---|
| α-Helix | β-Sheet | β-Turn | Random Coil | |
| Only HSA | 48.8 | 20.7 | 15.2 | 16.0 |
| HSA + 3-bromocarbazole (1:1) | 42.0 | 26.7 | 15.7 | 15.7 |
| HSA + 3-bromocarbazole (1:2) | 34.1 | 33.7 | 16.9 | 15.3 |
| HSA + 1,3,6,8-tetrabromocarbazole (1:1) | 41.8 | 26.8 | 14.9 | 16.6 |
| HSA + 1,3,6,8-tetrabromocarbazole (1:2) | 38.9 | 27.8 | 16.6 | 16.7 |
| Compound | Peaks | Peaks Position λex/λem (nm/nm) | Stokes Δλ (nm) | Intensity |
|---|---|---|---|---|
| Only HSA | Peak 1 | 280.0/348.0 | 68.0 | 619.2 |
| Peak 2 | 230.0/334.0 | 104.0 | 575.7 | |
| HSA-3-bromocarbazole | Peak 1 | 280.0/338.0 | 58.0 | 499.8 |
| Peak 2 | 230.0/328.0 | 98.0 | 505.5 | |
| HSA-1,3,6,8-tetrabromocarbazole | Peak 1 | 280.0/338.0 | 62.0 | 545.3 |
| Peak 2 | 230.0/330.0 | 100.0 | 553.2 |
| Compound | E (%) | J (cm3·L·mol−1) | R0 (nm) | r (nm) |
|---|---|---|---|---|
| HSA-3-bromocarbazole | 9.96 | 1.04×10−14 | 2.34 | 2.23 |
| HSA-1,3,6,8-tetrabromocarbazole | 7.55 | 0.90×10−14 | 2.28 | 2.27 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, X.; Yuan, D.; Pan, D. Interactions of Bromocarbazoles with Human Serum Albumin Using Spectroscopic Methods. Molecules 2018, 23, 3120. https://doi.org/10.3390/molecules23123120
Yan X, Yuan D, Pan D. Interactions of Bromocarbazoles with Human Serum Albumin Using Spectroscopic Methods. Molecules. 2018; 23(12):3120. https://doi.org/10.3390/molecules23123120
Chicago/Turabian StyleYan, Xiaodan, Dongjie Yuan, and Dandan Pan. 2018. "Interactions of Bromocarbazoles with Human Serum Albumin Using Spectroscopic Methods" Molecules 23, no. 12: 3120. https://doi.org/10.3390/molecules23123120




