Quantum Mechanical/Molecular Mechanical Analysis of the Catalytic Mechanism of Phosphoserine Phosphatase
Abstract
:1. Introduction
2. Materials and Methods
2.1. Active Site Model Calculations
2.2. MD Simulation
2.3. QM/MM Pathway Calculations
2.3.1. Minimum Energy Pathways and Potential Energy Scans
2.3.2. Umbrella Sampling Simulations
2.3.3. Analysis
3. Results
3.1. Enzymatic Reactant State Structure 1R
3.2. QM/MM Minimum Energy Pathways
3.2.1. Reaction Path of Step 1: Phosphate Transfer from Serine to Aspartate 11
3.2.2. Reaction Path of Step 2: Hydrolysis of Phosphoryl-Aspartate 11
3.2.3. Reaction Coordinates
3.3. Free Energy Calculations
3.3.1. One-Dimensional Reaction Profiles
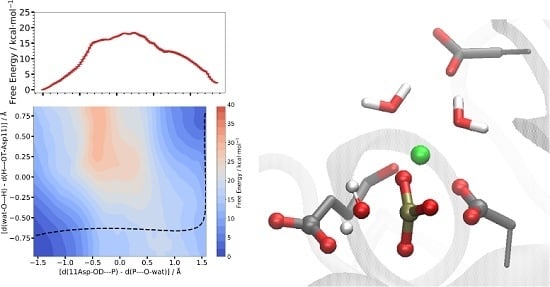
3.3.2. Two-Dimensional Reaction Profiles
Step 1
Step 2
4. Discussion
4.1. Phosphoryl Transfer Mechanism and the Nature of the Transition States
4.2. Choice of Reaction Coordinates
4.3. Enzymatic Efficiency of PSP
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PSPI | Phosphoserine phosphatase |
| PLS | phospho-l-serine |
| CPR | Conjugate Peak Refinement |
| MD | molecular dynamics |
| QM/MM | quantum mechanical/molecular mechanical |
| HAD | haloacid dehalogenase-like hydrolase |
| DFT | Density functional theory |
| B3LYP | Becke’s three parameter hybrid function |
| DFTB3/OB3 | Density functional tight-binding method with Third-Order Parametrization for Organic and |
| Biological Systems (3OB) | |
| SCC-DFTB | Self-consistent-charge density-functional tight-binding |
| SCF | self-consistend field |
| NPT | constant number of particles, N, pressure P, and temperature, T |
| NVT | constant number of particles, N, volume, V, and temperature, T |
| WHAM | Weighted Histogram Analyis Method |
| RC | Reaction coordinate |
References
- Dzeja, P.P.; Terzic, A. Phosphotransfer networks and cellular energetics. J. Exp. Biol. 2003, 206, 2039–2047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muzi-Falconi, M.; Brown, G.W.; Kelly, T.J. Controlling initiation during the cell cycle. DNA replication. Curr. Biol. 1996, 6, 229–233. [Google Scholar] [CrossRef]
- Dahmus, M.E. The role of multisite phosphorylation in the regulation of RNA polymerase II activity. Prog. Nucleic Acid Res. Mol. Biol. 1994, 48, 143–179. [Google Scholar] [PubMed]
- Sarre, T.F. The phosphorylation of eukaryotic initiation factor 2: A principle of translational control in mammalian cells. Biosystems 1989, 22, 311–325. [Google Scholar] [CrossRef]
- Cheung, W.L.; Ajiro, K.; Samejima, K.; Kloc, M.; Cheung, P.; Mizzen, C.A.; Beeser, A.; Etkin, L.D.; Chernoff, J.; Earnshaw, W.C.; et al. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell 2003, 113, 507–517. [Google Scholar] [CrossRef]
- Lad, C.; Williams, N.H.; Wolfenden, R. The rate of hydrolysis of phosphomonoester dianions and the exceptional catalytic proficiencies of protein and inositol phosphatases. Proc. Natl. Acad. Sci. USA 2003, 100, 5607–5610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hengge, A.C.; Sowa, G.A.; Wu, L.; Zhang, Z.Y. Nature of the transition state of the protein-tyrosine phosphatase-catalyzed reaction. Biochemistry 1995, 34, 13982–13987. [Google Scholar] [CrossRef] [PubMed]
- Warshel, A. Electrostatic origin of the catalytic power of enzymes and the role of preorganized active sites. J. Biol. Chem. 1998, 273, 27035–27038. [Google Scholar] [CrossRef] [PubMed]
- Choe, J.Y.; Iancu, C.V.; Fromm, H.J.; Honzatko, R.B. Metaphosphate in the active site of fructose-1,6- bisphosphatase. J. Biol. Chem. 2003, 278, 16015–16020. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Tatusov, R.L. Computer analysis of bacterial haloacid dehalogenases defines a large superfamily of hydrolases with diverse specificity. Application of an iterative approach to database search. J. Mol. Biol. 1994, 244, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Åqvist, J.; Kolmodin, K.; Florian, J.; Warshel, A. Mechanistic alternatives in phosphate monoester hydrolysis: What conclusions can be drawn from available experimental data? Chem. Biol. 1999, 6, R71–R80. [Google Scholar] [CrossRef]
- Catrina, I.E.; Hengge, A.C. Comparisons of phosphorothioate with phosphate transfer reactions for a monoester, diester, and triester: Isotope effect studies. J. Am. Chem. Soc. 2003, 125, 7546–7552. [Google Scholar] [CrossRef] [PubMed]
- Grzyska, P.K.; Czyryca, P.G.; Purcell, J.; Hengge, A.C. Transition state differences in hydrolysis reactions of alkyl versus aryl phosphate monoester monoanions. J. Am. Chem. Soc. 2003, 125, 13106–13111. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Lu, Z.; Jia, Y.; Dunaway-Mariano, D.; Herzberg, O. Dissociative phosphoryl transfer in PEP mutase catalysis: Structure of the enzyme/sulfopyruvate complex and kinetic properties of mutants. Biochemistry 2002, 41, 10270–10276. [Google Scholar] [CrossRef] [PubMed]
- Lahiri, S.D.; Zhang, G.; Dunaway-Mariano, D.; Allen, K.N. The pentacovalent phosphorus intermediate of a phosphoryl transfer reaction. Science 2003, 299, 2067–2071. [Google Scholar] [CrossRef] [PubMed]
- Moro-Furlani, A.M.; Turner, V.S.; Hopkinson, D.A. Genetical and biochemical studies on human phosphoserine phosphatase. Ann. Hum. Genet. 1980, 43, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Guynn, R.W.; Merrill, D.K.; Lund, K. The reactions of the phosphorylated pathway of l-serine biosynthesis: thermodynamic relationships in rat liver in vivo. Arch. Biochem. Biophys. 1986, 245, 204–211. [Google Scholar] [CrossRef]
- Dunlop, D.S.; Neidle, A. The origin and turnover of d-serine in brain. Biochem. Biophys. Res. Commun. 1997, 235, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Wolosker, H.; Sheth, K.N.; Takahashi, M.; Mothet, J.P.; Brady, R.O.; Ferris, C.D.; Snyder, S.H. Purification of serine racemase: Biosynthesis of the neuromodulator d-serine. Proc. Natl. Acad. Sci. USA 1999, 96, 721–725. [Google Scholar] [CrossRef] [PubMed]
- Wood, P.L. The co-agonist concept: Is the NMDA-associated glycine receptor saturated in vivo? Life Sci. 1995, 57, 301–310. [Google Scholar] [CrossRef]
- Matsui, T.; Sekiguchi, M.; Hashimoto, A.; Tomita, U.; Nishikawa, T.; Wada, K. Functional comparison of d-serine and glycine in rodents: The effect on cloned NMDA receptors and the extracellular concentration. J. Neurochem. 1995, 65, 454–458. [Google Scholar] [CrossRef] [PubMed]
- Peeraer, Y.; Rabijns, A.; Collet, J.; Schaftingen, E.V.; Ranter, C.D. How calcium inhibits the magnesium- dependent enzyme human phosphoserine phosphatase. Eur. J. Biochem. 2004, 271, 3421–3427. [Google Scholar] [CrossRef] [PubMed]
- Morais, M.C.; Zhang, W.; Baker, A.S.; Zhang, G.; Dunaway-Mariano, D.; Allen, K.N. The crystal structure of bacillus cereus phosphonoacetaldehyde hydrolase: Insight into catalysis of phosphorus bond cleavage and catalytic diversification within the HAD enzyme superfamily. Biochemistry 2000, 39, 10385–10396. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Mazurkie, A.S.; Dunaway-Mariano, D.; Allen, K.N. Kinetic evidence for a substrate-induced fit in phosphonoacetaldehyde hydrolase catalysis. Biochemistry 2002, 41, 13370–13377. [Google Scholar] [CrossRef] [PubMed]
- Allen, K.N.; Dunaway-Mariano, D. Phosphoryl group transfer: Evolution of a catalytic scaffold. Trends Biochem. Sci. 2004, 29, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Cho, H.S.; Kim, R.; Jancarik, J.; Yokota, H.; Nguyen, H.H.; Grigoriev, I.V.; Wemmer, D.E.; Kim, S.H. Structural characterization of the reaction pathway in phosphoserine phosphatase: Crystallographic “snapshots” of intermediate states. J. Mol. Biol. 2002, 319, 421–431. [Google Scholar] [CrossRef]
- Kim, H.Y.; Heo, Y.S.; Kim, J.H.; Park, M.H.; Moon, J.; Kim, E.; Kwon, D.; Yoon, J.; Shin, D.; Jeong, E.J.; et al. Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase. J. Biol. Chem. 2002, 277, 46651–46658. [Google Scholar] [CrossRef] [PubMed]
- Peeraer, Y.; Rabijns, A.; Verboven, C.; Collet, J.F.; Schaftingen, E.V.; Ranter, C.D. High-resolution structure of human phosphoserine phosphatase in open conformation. Acta Crystallogr. D Biol. Crystallogr. 2003, 59, 971–977. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Wang, W.; Kim, R.; Yokota, H.; Damo, S.; Kim, S.H.; Wemmer, D.; Kustu, S.; Yan, D. BeF acts as a phosphate analog in proteins phosphorylated on aspartate: Structure of a BeF complex with phosphoserine phosphatase. Proc. Natl. Acad. Sci. USA 2001, 98, 8525–8530. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Kim, R.; Jancarik, J.; Yokota, H.; Kim, S.H. Crystal structure of phosphoserine phosphatase from Methanococcus jannaschii, a hyperthermophile, at 1.8 Å resolution. Structure 2001, 9, 65–71. [Google Scholar] [CrossRef]
- Mildvan, A.S. Mechanisms of signaling and related enzymes. Proteins 1997, 29, 401–416. [Google Scholar] [CrossRef]
- Imhof, P.; Fischer, S.; Smith, J.C. Catalytic Mechanism of DNA Backbone Cleavage by the Restriction Enzyme EcoRV: A Quantum Mechanical/Molecular Mechanical Analysis. Biochemistry 2009, 48, 9061–9075. [Google Scholar] [CrossRef] [PubMed]
- Hou, G.; Zhu, X.; Elstner, M.; Cui, Q. A Modified QM/MM Hamiltonian with the Self-Consistent-Charge Density-Functional-Tight-Binding Theory for Highly Charged QM Regions. J. Chem. Theory Comput. 2012, 8, 4293–4304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grigorenko, B.L.; Rogov, A.V.; Nemukhin, A.V. Mechanism of triphosphate hydrolysis in aqueous solution: QM/MM simulations in water clusters. J. Phys. Chem. B 2006, 110, 4407–4412. [Google Scholar] [CrossRef] [PubMed]
- Vivo, M.D.; Cavalli, A.; Carloni, P.; Recanatini, M. Computational study of the phosphoryl transfer catalyzed by a cyclin-dependent kinase. Chemistry 2007, 13, 8437–8444. [Google Scholar] [CrossRef] [PubMed]
- Duarte, F.; Barrozo, A.; Åqvist, J.; Williams, N.H.; Kamerlin, S.C.L. The Competing Mechanisms of Phosphate Monoester Dianion Hydrolysis. J. Am. Chem. Soc. 2016, 138, 10664–10673. [Google Scholar] [CrossRef] [PubMed]
- Korhonen, H.; Mikkola, S.; Williams, N.H. The mechanism of cleavage and isomerisation of RNA promoted by an efficient dinuclear Zn2+ complex. Chem. A Eur. J. 2012, 18, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Golden, B.L. Two distinct catalytic strategies in the hepatitis delta virus ribozyme cleavage reaction. Biochemistry 2011, 50, 9424–9433. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, J.C.; Strobel, S.A. Catalytic strategies of self-cleaving ribozymes. Acc. Chem. Res. 2008, 41, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Florián, J.; Åqvist, J.; Warshel, A. On the Reactivity of Phosphate Monoester Dianions in Aqueous Solution: Bronsted Linear Free-Energy Relationships Do Not Have an Unique Mechanistic Interpretation. J. Am. Chem. Soc. 1998, 120, 11524–11525. [Google Scholar] [CrossRef]
- Florián, J.; Strajbl, M.; Warshel, A. Conformational flexibility of phosphate, phosphonate, and phosphorothioate methyl esters in aqueous solution. J. Am. Chem. Soc. 1998, 120, 7959–7966. [Google Scholar] [CrossRef]
- Florián, J.; Warshel, A. Phosphate ester hydrolysis in aqueous solution: Associative versus dissociative mechanisms. J. Phys. Chem. B 1998, 102, 719–734. [Google Scholar] [CrossRef]
- Hu, C.H.; Brinck, T. Theoretical Studies of the Hydrolysis of the Methyl Phosphate Anion. J. Phys. Chem. A 1999, 103, 5379–5386. [Google Scholar] [CrossRef]
- Humphry, T.; Forconi, M.; Williams, N.H.; Hengge, A.C. An Altered Mechanism of Hydrolysis for a Metal-Complexed Phosphate Diester. J. Am. Chem. Soc. 2002, 124, 14860–14861. [Google Scholar] [CrossRef] [PubMed]
- Mercero, J.M.; Barrett, P.; Lam, C.W.; Fowler, J.E.; Ugalde, J.M.; Pedersen, L.G. Quantum Mechanical Calculations on Phosphate Hydrolysis Reactions. J. Comput. Chem. 2000, 21, 43–51. [Google Scholar] [CrossRef]
- Iche-Tarrat, N.; Barthelat, J.C.; Rinaldi, D.; Vigroux, A. Theoretical Studies of the Hydroxide-Catalyzed P-O Cleavage Reactions of Neutral Phosphate Triesters and Diesters in Aqueous Solution: Examination of the Changes Induced by H/Me Substitution. J. Phys. Chem. B 2005, 109, 22570–22580. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, P.J.; Herschlag, D. Alkaline Phosphatase Revisited: Hydrolysis of Alkyl Phosphates. Biochemistry 2001, 41, 3207–3225. [Google Scholar] [CrossRef] [PubMed]
- James Borden, D.C.C.; Florian, J. Transition State Analogues for Nucleotidyl Transfer Reactions: Structure and Stability of Pentavalent Vanadate and Phosphate Ester Dianions. J. Phys. Chem. B 2005, 110, 14988–14999. [Google Scholar] [CrossRef] [PubMed]
- Peck, A.; Sunden, F.; Andrews, L.D.; Pande, V.S.; Herschlag, D. Tungstate as a Transition State Analog for Catalysis by Alkaline Phosphatase. J. Mol. Biol. 2016, 428, 2758–2768. [Google Scholar] [CrossRef] [PubMed]
- Emmanuel, Y.; Tirel, N.H.W. Enhancing Phosphate Diester Cleavage by a Zinc Complex through Controlling Nucleophile Coordination. Chem. A Eur. J. 2015, 21, 7053–7056. [Google Scholar] [Green Version]
- Mones, L.; Kulhanek, P.; Florian, J.; Istvan, S.; Fuxreiter, M. Probing the Two-Metal Ion Mechanism in the Restriction Endonuclease BamHI. Biochemistry 2007, 46, 14514–14523. [Google Scholar] [CrossRef] [PubMed]
- Boero, M.; Tateno, M.; Terakura, K.; Oshiyama, A. Double-Metal-Ion/Single-Metal-Ion Mechanisms of the Cleavage Reaction of Ribozymes: First-Principles Molecular Dynamics Simulations of a Fully Hydrated Model System. J. Chem. Theory Comput. 2005, 1, 925–934. [Google Scholar] [CrossRef] [PubMed]
- Re, S.; Jung, J.; Ten.no, S.; Sugita, Y. A two-dimensional energy surface of the phosphoryl transfer reaction catalyzed by phosphoserine phosphatase. Chem. Phys. Lett. 2009, 480, 284–288. [Google Scholar] [CrossRef]
- Ahlrichs, R.; Bar, M.; Haser, M.; Horn, H.; Kolmel, C. Electronic Structure Calculations on Workstation Computers: The Program System TURBOMOLE. Chem. Phys. Lett. 1989, 162, 165–169. [Google Scholar] [CrossRef]
- Cui, Q.; Elstner, M.; Kaxiras, E.; Frauenheim, T.; Karplus, M. A QM/MM Implementation of the Self-Consistent Charge Density Functional Tight Binding (SCC-DFTB) Method. J. Phys. Chem. B 2001, 105, 569–585. [Google Scholar] [CrossRef]
- Gaus, M.; Lu, X.; Elstner, M.; Cui, Q. Parameterization of DFTB3/3OB for Sulfur and Phosphorus for Chemical and Biological Applications. J. Chem. Theory Comput. 2014, 10, 1518–1537. [Google Scholar] [CrossRef] [PubMed]
- Batebi, H.; Imhof, P. Phosphodiester hydrolysis computed for cluster models of enzymatic active sites. Theor. Chem. Acc. 2016, 135, 262. [Google Scholar] [CrossRef]
- Hou, G.; Cui, Q. Stabilization of Different Types of Transition States in a Single Enzyme Active Site: QM/MM Analysis of Enzymes in the Alkaline Phosphatase Superfamily. J. Am. Chem. Soc. 2013, 135, 10457–10469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brooks, B.R.; Brooks, C.L.; Mackerell, A.D.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, S.; Karplus, M. Conjugate Peak Refinement: An algorithm for finding reaction paths and accurate transition states in systems with many degrees of freedom. Chem. Phys. Lett. 1992, 194, 252–261. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Essman, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Feller, S.E.; Zhang, Y.; Pastor, R.W.; Brooks, B.R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995, 103, 4613–4621. [Google Scholar] [CrossRef]
- Kalibaeva, G.; Ferrario, M.; Ciccotti, G. Constant pressure-constant temperature molecular dynamics: A correct constrained NPT ensemble using the molecular virial. Mol. Phys. 2003, 101, 765–778. [Google Scholar] [CrossRef]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comp. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2007, 31, 671–690. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Mark, P.; Nilsson, L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Field, M.J.; Bash, P.A.; Karplus, M. A combined quantum mechanical and molecular mechanical potential for molecular dynamics simulations. J. Comput. Chem. 1990, 11, 700–733. [Google Scholar] [CrossRef]
- Braga, C.; Travis, K.P. A configurational temperature Nosé-Hoover thermostat. J. Chem. Phys. 2005, 123, 134101. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Rosenberg, J.M.; Bouzida, D.; Swendsen, R.H.; Kollman, P.A. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 1992, 13, 1011–1021. [Google Scholar] [CrossRef]
- Souaille, M.; Roux, B. Extension to the weighted histogram analysis method: Combining umbrella sampling with free energy calculations. Comput. Phys. Commun. 2001, 135, 40–57. [Google Scholar] [CrossRef]
- Dijkstra, E.A. A note on two problems in connection with graphs. Numer. Math. 1959, 1, 269. [Google Scholar] [CrossRef]
- Sabato, G.D.; Jencks, W.P. Mechanism and Catalysis of Reactions of Acyl Phosphates. II. Hydrolysis. J. Am. Chem. Soc. 1961, 83, 4400–4405. [Google Scholar] [CrossRef]
- Admiraal, S.J.; Schneider, B.; Meyer, P.; Janin, J.; Veron, M.; Deville-Bonne, D.; Herschlag, D. Nucleophilic activation by positioning in phosphoryl transfer catalyzed by nucleoside diphosphate kinase. Biochemistry 1999, 38, 4701–4711. [Google Scholar] [CrossRef] [PubMed]
- Laio, A.; Parrinello, M. Escaping free-energy minima. Proc. Natl. Acad. Sci. USA 2002, 99, 12562–12566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barducci, A.; Bussi, G.; Parrinello, M. Well-tempered metadynamics: A smoothly converging and tunable free-energy method. Phys. Rev. Lett. 2008, 100, 020603. [Google Scholar] [CrossRef] [PubMed]
- Dama, J.; Parrinello, M.; Voth, G. Well-Tempered Metadynamics Converges Asymptotically. Phys. Rev. Lett. 2014, 112, 240602. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Sode, O.; Dama, J.F.; Voth, G.A. Simulating Protein Mediated Hydrolysis of ATP and Other Nucleoside Triphosphates by Combining QM/MM Molecular Dynamics with Advances in Metadynamics. J. Chem. Theory Comput. 2017, 13, 2332–2341. [Google Scholar] [CrossRef] [PubMed]
- Weinan, E.; Weiqing, R.; Vanden-Eijnden, E. String method for the study of rare events. Phys. Rev. B 2002, 66, 052301. [Google Scholar]
- Rosta, E.; Nowotny, M.; Yang, W.; Hummer, G. Catalytic Mechanism of RNA Backbone Cleavage by Ribonuclease H from QM/MM Simulations. J. Am. Chem. Soc. 2011, 133, 8934–8941. [Google Scholar] [CrossRef] [PubMed]
- Newman, J.W.; Morisseau, C.; Harris, T.R.; Hammock, B.D. The soluble epoxide hydrolase encoded by EPXH2 is a bifunctional enzyme with novel lipid phosphate phosphatase activity. Proc. Natl. Acad. Sci. USA 2003, 100, 1558–1563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vivo, M.D.; Ensing, B.; Peraro, M.D.; Gomez, G.A.; Christianson, D.W.; Klein, M.L. Proton shuttles and phosphatase activity in soluble epoxide hydrolase. J. Am. Chem. Soc. 2007, 129, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Fersht, A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding; W.H.Freeman & Co.: New York, NY, USA, 1999. [Google Scholar]
- Collet, J.F.; Stroobant, V.; Schaftingen, E.V. Mechanistic studies of phosphoserine phosphatase, an enzyme related to P-type ATPases. J. Biol. Chem. 1999, 274, 33985–33990. [Google Scholar] [CrossRef] [PubMed]











| Step 1 | Step 2 | |||
|---|---|---|---|---|
| TS | Prod | TS | Prod | |
| CPR | 1.6 | −14.3 | 29.8 | 0.1 |
| 1D-Scan, OPO | 10.7 | −9.5 | 22.8 | −3.9 |
| 1D-US, OPO | 8.2 ± 0.1 | −2.0 ± 0.0 | 18.1 ± 0.2 | −2.6 ± 0.0 |
| 2D-Scan, OP, PO | 12.7 | −9.9 | 23.3 | 14.1 |
| 2D-US, OP, PO | 7.8 ± 0.1 | −5.7 ± 0.1 | 21.5 ± 0.1 | 1.1 ± 0.1 |
| 2D-Scan, OPO, OHO | 8.2 | −12.0 | 36.9 | 11.9 |
| 2D-US, OPO, OHO | 5.4 ± 0.1 | −6.8 ± 0.5 | 18.4 ± 0.1 | 2.3 ± 0.1 |
| RC | Ser-OG–P | P–O-Asp11 | H–OD-Asp13 | Ser-OG–H | Ser-OT–H | ||
|---|---|---|---|---|---|---|---|
| CPR | IM | 0.73 | 1.74 | 2.81 | 1.01 | 1.64 | 2.56 |
| TS | 0.76 | 1.83 | 2.71 | 1.23 | 1.20 | 2.74 | |
| P | 0.91 | 2.76 | 1.78 | 1.79 | 0.99 | 3.13 | |
| 1D Scan | IM | −1.5 | 1.72 | 3.25 | 1.03 | 2.44 | 1.54 |
| OPO | TS | −0.9 | 2.04 | 2.94 | 1.60 | 1.02 | 2.84 |
| P | 1.2 | 3.09 | 1.87 | 1.76 | 0.99 | 3.52 | |
| 1D US | IM | −1.3 | 1.66 ± 0.03 | 2.78 ± 0.05 | 1.84 ± 0.25 | 2.46 ± 0.13 | 1.00 ± 0.03 |
| OPO | TS | −0.3 | 2.36 ± 0.09 | 2.54 ± 0.08 | 1.71 ± 0.13 | 1.01 ± 0.03 | 3.02 ± 0.18 |
| P | 1.0 | 2.71 ± 0.06 | 1.80 ± 0.04 | 1.83 ± 0.13 | 0.99 ± 0.02 | 3.03±0.18 | |
| 2D-Scan | IM | 1.7, 2.9 | 1.69 | 2.90 | 1.05 | 2.52 | 1.48 |
| OP, | TS | 1.8, 2.2 | 1.83 | 2.22 | 1.60 | 1.03 | 2.57 |
| PO | P | 2.8, 1.8 | 2.80 | 1.79 | 1.78 | 0.99 | 3.18 |
| 2D-US | IM | 1.7, 2.7 | 1.68 ± 0.03 | 2.72 ± 0.05 | 1.89 ± 0.31 | 2.44 ± 0.12 | 1.00 ± 0.03 |
| OP, | TS | 2.2, 2.5 | 2.22 ± 0.06 | 2.50 ± 0.05 | 1.68 ± 0.11 | 1.01 ± 0.03 | 2.87 ± 0.14 |
| PO | P | 2.7, 1.8 | 2.71 ± 0.05 | 1.80 ± 0.04 | 1.83 ± 0.14 | 0.99 ± 0.03 | 3.03 ± 0.17 |
| 2D-Scan | IM | −1.5, −0.7 | 1.74 | 3.24 | 1.00 | 1.70 | 2.38 |
| OPO, | TS | −1.3, 0.0 | 1.84 | 3.14 | 1.31 | 1.31 | 2.77 |
| OHO | P | 1.2, 0.7 | 3.09 | 1.87 | 1.75 | 0.97 | 3.50 |
| 2D-US | IM | 1.1, −0.8 | 1.74 ± 0.04 | 2.76 ± 0.05 | 1.01 ± 0.03 | 1.71 ± 0.06 | 2.46 ± 0.26 |
| OPO, | TS | −0.6, 0.3 | 1.94 ± 0.05 | 2.63 ± 0.06 | 1.54 ± 0.04 | 1.04 ± 0.02 | 2.72 ± 0.13 |
| OHO | P | 1.0, 0.8 | 2.78 ± 0.06 | 1.78 ± 0.04 | 1.79 ± 0.05 | 0.97 ± 0.02 | 3.11 ± 0.17 |
| RC | P–O-Asp11 | Wat-O–P | H–OD-Asp13 | Wat-O–H | H–OT-Asp11 | ||
|---|---|---|---|---|---|---|---|
| CPR | IM | 0.26 | 1.74 | 3.82 | 3.50 | 0.99 | 4.00 |
| TS | 0.52 | 2.51 | 2.14 | 1.52 | 1.03 | 2.47 | |
| P | 0.64 | 3.04 | 1.66 | 1.12 | 2.93 | 1.28 | |
| 1D Scan | IM | −1.5 | 1.75 | 3.25 | 2.06 | 0.97 | 2.88 |
| OPO | TS | 1.1 | 3.04 | 1.95 | 1.57 | 1.02 | 2.38 |
| P | 1.6 | 3.30 | 1.68 | 1.52 | 2.90 | 1.02 | |
| 1D US | IM | −1.6 | 1.78 ± 0.05 | 3.40 ± 0.07 | 3.44 ± 0.64 | 0.98 ± 0.03 | 3.18 ± 0.31 |
| OPO | TS | 0.5 | 3.26 ± 0.10 | 2.92 ± 0.10 | 3.87 ± 0.61 | 0.97 ± 0.02 | 2.65 ± 0.38 |
| P | 1.6 | 3.26 ± 0.06 | 1.61 ± 0.03 | 4.33 ± 0.26 | 3.43 ± 0.08 | 1.01 ± 0.03 | |
| 2D-Scan | IM | 1.8, 3.8 | 1.79 | 3.78 | 1.73 | 0.99 | 3.81 |
| OP, | TS | 2.6, 2.1 | 2.60 | 2.20 | 1.61 | 1.01 | 2.62 |
| PO | P | 3.2, 1.8 | 3.20 | 1.78 | 0.99 | 1.77 | 2.34 |
| 2D-US | IM | 1.8, 3.9 | 1.76 ± 0.04 | 3.61 ± 0.05 | 4.17 ± 0.35 | 0.98 ± 0.03 | 3.33 ± 0.30 |
| OP, | TS | 2.9, 2.3 | 2.94 ± 0.06 | 1.82 ± 0.04 | 3.16 ± 0.13 | 2.96 ± 0.23 | 1.35 ± 0.21 |
| PO | P | 3.1, 1.7 | 3.15 ± 0.05 | 1.65 ± 0.03 | 4.58 ± 0.53 | 3.42 ± 0.14 | 1.01 ± 0.05 |
| 2D-Scan | IM | −1.5, −1.0 | 1.75 | 3.25 | 2.83 | 0.96 | 1.95 |
| OPO, | TS | 1.4, −0.3 | 3.20 | 1.76 | 1.15 | 1.35 | 1.62 |
| OHO | P | 1.5, 1.0 | 3.24 | 1.73 | 1.66 | 1.97 | 0.98 |
| 2D-US | IM | −1.6, −0.8 | 1.77 ± 0.04 | 3.40 ± 0.07 | 1.72 ± 0.05 | 1.00 ± 0.03 | 3.41 ± 0.20 |
| OPO, | TS | -0.5, -0.6 | 2.85 ± 0.06 | 1.67 ± 0.04 | 1.62 ± 0.06 | 1.01 ± 0.03 | 3.19 ± 0.37 |
| OHO | P | 1.6, 0.8 | 3.29 ± 0.06 | 1.68 ± 0.04 | 1.00 ± 0.02 | 1.74 ± 0.05 | 2.43 ± 0.26 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krachtus, D.; Smith, J.C.; Imhof, P. Quantum Mechanical/Molecular Mechanical Analysis of the Catalytic Mechanism of Phosphoserine Phosphatase. Molecules 2018, 23, 3342. https://doi.org/10.3390/molecules23123342
Krachtus D, Smith JC, Imhof P. Quantum Mechanical/Molecular Mechanical Analysis of the Catalytic Mechanism of Phosphoserine Phosphatase. Molecules. 2018; 23(12):3342. https://doi.org/10.3390/molecules23123342
Chicago/Turabian StyleKrachtus, Dieter, Jeremy C. Smith, and Petra Imhof. 2018. "Quantum Mechanical/Molecular Mechanical Analysis of the Catalytic Mechanism of Phosphoserine Phosphatase" Molecules 23, no. 12: 3342. https://doi.org/10.3390/molecules23123342