Search for Fibrous Aggregates Potentially Useful in Regenerative Medicine Formed under Physiological Conditions by Self-Assembling Short Peptides Containing Two Identical Aromatic Amino Acid Residues
Abstract
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. General Information
3.2. Peptide Synthesis
Manual Solid-Phase Peptide Synthesis (SPPS)
3.3. Aggregation Studies
3.3.1. Spectroscopic Measurements with Congo Red
3.3.2. Spectroscopic Measurements with Thioflavin T
3.3.3. Peptide Analysis under Polarized Light
3.4. Surface Properties of Peptides
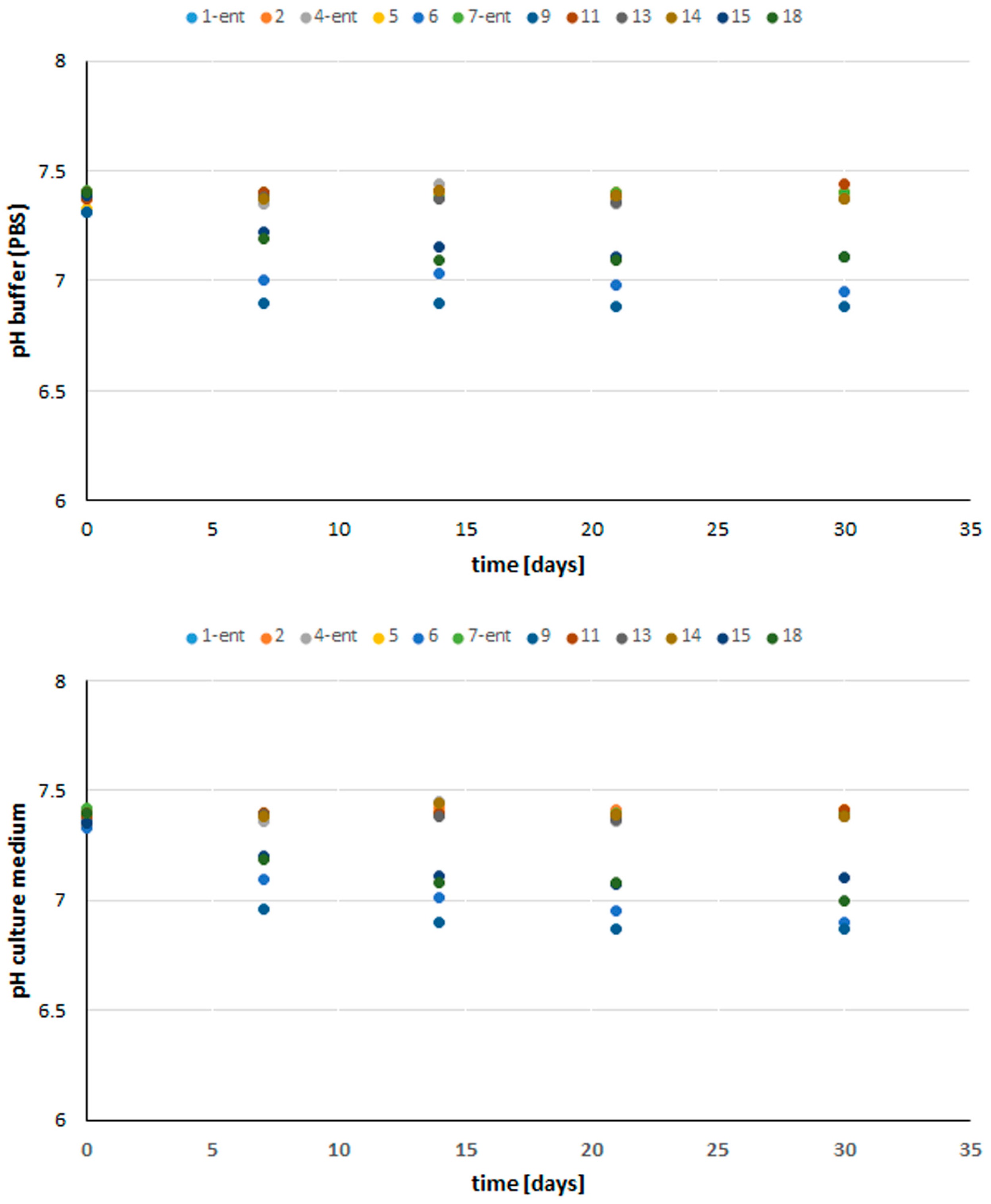
3.5. Stability and Biological Properties of Peptide Materials
3.5.1. Stability of Peptides under In Vitro Conditions
3.5.2. Cell Cytotoxicity Test
3.5.3. Cytotoxicity Test
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rajagopal, K.; Schneider, J.P. Self-assembling peptides and proteins for nanotechnological applications. Curr. Opin. Struct. Biol. 2004, 14, 480–486. [Google Scholar] [CrossRef] [PubMed]
- Ulijn, R.V. Molecular Self-assembly: Best of Both Worlds. Nat. Nanotechnol. 2015, 10, 295–296. [Google Scholar] [CrossRef] [PubMed]
- Lampel, A.; McPhee, S.A.; Park, H.-A.; Scott, G.G.; Humagain, S.; Hekstra, D.R.; Yoo, B.; Frederix, P.W.J.M.; Li, T.-D.; Abzalimov, R.R.; et al. Polymeric Peptide Pigments with Sequence-encoded Properties. Science 2017, 356, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Pappas, C.G.; Shafi, R.; Sasselli, I.R.; Siccardi, H.; Wang, T.; Narang, V.; Abzalimov, R.; Wijerathne, N.; Ulijn, R.V. Dynamic Peptide Libraries for the Discovery of Supramolecular Nanomaterials. Nat. Nanotechnol. 2016, 11, 960–967. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Xu, Q.; Dong, C.; Lee, S.S.; Gao, L.; Li, Y.; D’Ortenzio, M.; Wu, J. Self-Assembling Peptide Nanofibrous Hydrogel as a Versatile Drug Delivery Platform. Curr. Pharm. Des. 2015, 21, 4342–4354. [Google Scholar] [CrossRef] [PubMed]
- Sundar, S.; Chen, Y.; Tong, Y.W. Delivery of Therapeutics and Molecules Using Self-Assembled Peptides. Curr. Med. Chem. 2014, 21, 2469–2479. [Google Scholar] [CrossRef] [PubMed]
- Kalafatovic, D.; Nobis, M.; Son, J.; Anderson, K.I.; Ulijn, R.V. MMP-9 Triggered Self-assembly of Doxorubicin Nanofiber Depots Halts Tumor Growth. Biomaterials 2016, 98, 192–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadatmousavi, P.; Soltani, M.; Nazarian, R.; Jafari, M.; Chen, P. Self-Assembling Peptides: Potential Role in Tumor Targeting. Curr. Pharm. Biotechnol. 2011, 12, 1089–1100. [Google Scholar] [CrossRef]
- Sharma, P.P.; Rathi, B.; Rodrigues, J.; Gorobets, N.Y. Self-Assembled Peptide Nanoarchitectures: Applications and Future Aspects. Curr. Top. Med. Chem. 2015, 15, 1268–1289. [Google Scholar] [CrossRef] [PubMed]
- Nune, M.; Kumaraswamy, P.; Krishnan, U.M.; Sethuraman, S. Peptide Nanofibers for Functional Tissue Regeneration. Curr. Prot. Pept. Sci. 2013, 14, 70–84. [Google Scholar] [CrossRef]
- Yu, C.Y.; Huang, W.; Li, Z.P.; Lei, X.Y.; He, D.X.; Sun, L. Progress in Self-assembling Peptide-based Nanomaterials for Biomedical Applications. Curr. Top. Med. Chem. 2016, 16, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Anderson, H.J.; Sahoo, J.K.; Ulijn, R.V.; Dalby, M.J. Mesenchymal Stem Cell Fate: Applying Biomaterials for Control of Stem Cell Behavior. Front. Bioeng. Biotechnol. 2016, 4, a38. [Google Scholar] [CrossRef] [PubMed]
- Dehsorkhi, A.; Castelletto, V.; Hamley, I.W. Self-assembling amphiphilic peptides. J. Pept. Sci. 2014, 20, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Rubert Pérez, C.M.; Stephanopoulos, N.; Sur, S.; Lee, S.S.; Newcomb, C.; Stupp, S.I. The Powerful Functions of Peptide-Based Bioactive Matrices for Regenerative Medicine. Ann. Biomed. Eng. 2015, 43, 501–514. [Google Scholar] [CrossRef] [PubMed]
- Hosseinkhani, H.; Hong, P.D.; Yu, D.S. Self-Assembled Proteins and Peptides for Regenerative Medicine. Chem. Rev. 2013, 113, 4837–4861. [Google Scholar] [CrossRef] [PubMed]
- Holmes, T.C.; de Lacalle, S.; Su, X.; Liu, G.; Rich, A.; Zhang, S. Extensive neurite outgrowth and active synapse formation on selfassembling peptide scaffolds. Proc. Natl. Acad. Sci. USA. 2000, 97, 6728–6733. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Leung, K.K.G.; Huanxing, S.; Yuan, Q.; Wang, Li.; TakHo Chu, T.H.; Zhang, W.; Pu, J.K.S.; Ng, G.K.P.; Wong, W.M.; et al. Self-assembling peptide nanofiber scaffold promotes the reconstruction of acutely injured brain. Nanomed. Nanotechnol. Biol. Med. 2009, 5, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Nagrath, D.; Chen, P.C.; Berthiaume, F.; Yarmush, M.L. Three-dimensional primary hepatocyte culture in synthetic self-assembling peptide hydrogel. Tissue Eng. Part A 2008, 14, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Ellis-Behnke, R.G.; Liang, Y.-X.; You, S.-W.; Tay, D.K.C.; Zhang, S.; So, K.-F. Nano neuro knitting: Peptide nanofiber scaffold for brain repair and axon regeneration with functional return of vision. Proc. Natl. Acad. Sci. USA. 2006, 103, 5054–5059. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Shi, G.-S.; Ren, L.-F.; Hu, F.-Q.; Li, S.-L.; Xie, Z.-J. Designer self-assembling peptide scaffold stimulates pre-osteoblast attachment, spreading and proliferation. J. Mater. Sci. Mater. Med. 2009, 20, 1475–1481. [Google Scholar] [CrossRef] [PubMed]
- Gilead, S.; Gazit, E. Self-Organization of Short Peptide Fragments: From Amyloid Fibrils to Nanoscale Supramolecular Assemblies. Supramol. Chem. 2005, 17, 87–92. [Google Scholar] [CrossRef]
- Gazit, E. The “correctly-folded” state of proteins: Is it a metastable state? Angew. Chem. Int. Ed. 2002, 41, 257–259. [Google Scholar] [CrossRef]
- Knowles, T.P.J.; Buehler, M.J. Nanomechanics of functional and pathological amyloid materials. Nat. Nanotechnol. 2011, 6, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Gazit, E. A possible role for pi-stacking in the self-assembly of amyloid fibrils. FASEB J. 2002, 16, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Gazit, E. Self-assembly of short aromatic peptides: From amyloid disease to nanotechnology. NanoBioTechnology 2005, 1, 286–288. [Google Scholar] [CrossRef]
- Wang, W.; Yang, Z.; Patanavanich, S.; Xu, B.; Chau, Y. Controlling self-assembly within nanospace for peptide nanoparticle fabrication. Soft Matter 2008, 4, 1617–1620. [Google Scholar] [CrossRef]
- Mankar, S.; Anoop, A.; Sen, S.; Maji, S.K. Nanomaterials: Amyloids reflect their brighter side. Nano Rev. 2011, 2, 6032. [Google Scholar] [CrossRef] [PubMed]
- Marchesan, S.; Vargiu, A.V.; Styan, K.E. The Phe-Phe Motif for Peptide Self-Assembly in Nanomedicine. Molecules 2015, 20, 19775–19788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Chau, Y. Self-assembled peptide nanorods as building blocks of fractal patterns. Soft Matter 2009, 5, 4893–4898. [Google Scholar] [CrossRef]
- Gazit, E. Molecular self-assembly: Searching sequence space. Nat. Chem. 2015, 7, 14–15. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, S.; Albericio, F.; Kros, A. Amphiphilic peptides and their cross-disciplinary role as building blocks for nanoscience. Chem. Soc. Rev. 2010, 39, 241–263. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanan, A.; Zhang, S.; Hauser, C.A. Short self-assembling peptides as building blocks for modern nanodevices. Trends Biotechnol. 2012, 30, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Reches, M.; Gazit, E. Controlled patterning of aligned self-assembled peptide nanotubes. Nat. Nanotechnol. 2006, 1, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Gazit, E. Self-assembled peptide nanostructures: The design of molecular building blocks and their technological utilization. Chem. Soc. Rev. 2007, 36, 1263–1269. [Google Scholar] [CrossRef] [PubMed]
- Colombo, G.; Soto, P.; Gazit, E. Peptide self-assembly at the nanoscale: A challenging target for computational and experimental biotechnology. Trends Biotechnol. 2007, 25, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Kol, N.; Adler-Abramovich, L.; Barlam, D.; Shneck, R.Z.; Gazit, E.; Rousso, I. Self-assembled peptide nanotubes are uniquely rigid bioinspired supramolecular structures. Nano Lett. 2005, 5, 1343–1346. [Google Scholar] [CrossRef] [PubMed]
- Reches, M.; Gazit, E. Casting metal nanowires within discrete self-assembled peptide nanotubes. Science 2003, 300, 625–627. [Google Scholar] [CrossRef] [PubMed]
- Reches, M.; Gazit, E. Formation of Closed-Cage Nanostructures by Self-Assembly of Aromatic Dipeptides. Nano Lett. 2004, 4, 581–585. [Google Scholar] [CrossRef]
- Kolesinska, B.; Rozniakowski, K.K.; Fraczyk, J.; Relich, I.; Papini, A.M.; Kamiński, Z.J. The effect of counterion and tertiary amine on the efficiency of N-triazinylammonium sulfonates in solution and solid-phase peptide synthesis. Eur. J. Org. Chem. 2015, 2, 401–408. [Google Scholar] [CrossRef]
- Klunk, W.E.; Pettegrew, J.W.; Abraham, D.J. Quantitative evaluation of congo red binding to amyloid-like proteins with a beta-pleated sheet conformation. J. Histochem. Cytochem. 1989, 37, 1273–1281. [Google Scholar] [CrossRef] [PubMed]
- Klunk, W.E.; Jacob, R.F.; Mason, R.P. Quantifying amyloid by congo red spectral shift assay. Methods Enzymol. 1999, 309, 285–305. [Google Scholar] [PubMed]
- Klunk, W.E.; Jacob, R.F.; Mason, R.P. Quantifying amyloid beta-peptide (Abeta) aggregation using the Congo red-Abeta (CR-abeta) spectrophotometric assay. Anal. Biochem. J. 1999, 266, 66–76. [Google Scholar] [CrossRef] [PubMed]
- O’Nuallain, B.; Shivaprasad, S.; Kheterpal, I.; Wetzel, R. Thermodynamics of A beta(1–40) amyloid fibril elongation. Biochemistry 2005, 44, 12709–12718. [Google Scholar] [CrossRef] [PubMed]
- Shivaprasad, S.; Wetzel, R. Scanning cysteine mutagenesis analysis of A beta-(1-40) amyloid fibrils. J. Biol. Chem. 2006, 281, 993–1000. [Google Scholar] [CrossRef] [PubMed]
- Groenning, M. Binding mode of Thioflavin T and other molecular probes in the context of amyloid fibrils-current status. J. Chem. Biol. 2010, 3, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Reinke, A.A.; Gestwicki, J.E. Insight into Amyloid Structure Using Chemical Probes. Chem. Biol. Drug Des. 2011, 77, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Swiontek, M.; Rozniakowski, K.; Fraczyk, J.; Lipinski, W.; Galecki, K.; Wysocki, S.; Dupont, B.G.R.; Kaminski, Z.J.; Kolesińska, B. The quest for the shortest fragments of A (13–19) and B (12–17) responsible for the aggregation of human insulin. Nanomedicine 2016, 11, 2083–2101. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [PubMed]
- Dobson, C.M. Protein folding and misfolding. Nature 2003, 426, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Swiontek, M.; Kaminski, Z.J.; Kolesinska, B.; Seebach, D. Visible-Light Microscopic Discovery of up to 150 μm Long Helical Amyloid Fibrils built of the Dodecapeptide H-(Val-Ala-Leu)4-OH and of Decapeptides Derived from Insulin. Chem. Biodivers. 2016, 13, 1111–1117. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.Y.; Shaughnessy, M.C.; Zho, Z.; Noh, H.; Vogler, E.A.; Donahuea, H.J. Surface energy effects on osteoblast spatial growth and mineralization. Biomaterials 2008, 29, 1776–1784. [Google Scholar] [CrossRef] [PubMed]
- Kubies, D.; Himmlová, L.; Riedel, T.; Chánová, E.; Balík, K.; Douděrová, M.; Bártová, J.; Pešáková, V. The interaction of osteoblasts with bone-implant materials: 1. The effect of physicochemical surface properties of implant materials. Physiol. Res. 2011, 60, 95–111. [Google Scholar] [PubMed]
- Bogun, M.; Stodolak, E.; Menaszek, E.; Scisłowska-Czarnecka, A. Composites Based on Poly-e-Caprolactone and Calcium Alginate Fibres Containing Ceramic Nanoadditives for Use in Regenerative Medicine. FIBRES TEXTILES East. Eur. 2011, 19, 17–21. [Google Scholar]
- Massimo, S. Biochemical and biophysical features of both oligomer⁄fibril and cell membrane in amyloid cytotoxicity. FEBS J. 2010, 277, 4602–4613. [Google Scholar]
- Xue, W.-F.; Hellewell, A.L.; Gosal, W.S.; Homans, S.W.; Hewitt, E.W.; Radford, S.E. Fibril Fragmentation Enhances Amyloid Cytotoxicity. J. Biol. Chem. 2009, 284, 34272–34282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lidington, E.A.; Moyes, D.L.; McCormack, A.M.; Rose, M.L. A comparison of primary endothelial cells and endothelial cell lines for studies of immune interactions. Transpl. Immunol. 1999, 7, 239–246. [Google Scholar] [CrossRef]
- Ho, M.-H.; Guo, Z.-M.; Chunga, J.; Goodwin, J.S.; Xie, H. Characterization of Innate Immune Responses of Human Endothelial Cells Induced by Porphyromonas gingivalis and Their Derived Outer Membrane Vesicles. Front. Cell Infect. Microbiol. 2016, 6, 139. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, E.; Colescott, R.L.; Bossinger, C.D.; Cook, P.I. Color test for detection of free terminal amino groups in the solid-phase synthesis of peptides. Anal. Biochem. 1970, 34, 595–598. [Google Scholar] [CrossRef]
- Edgell, C.J.; McDonald, C.C.; Graham, J.B. Permanent cell line expressing human factor VIII-related antigen established by hybridization. Proc. Natl. Acad. Sci. USA 1983, 80, 3734–3737. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds 1–18 are available from the authors. |
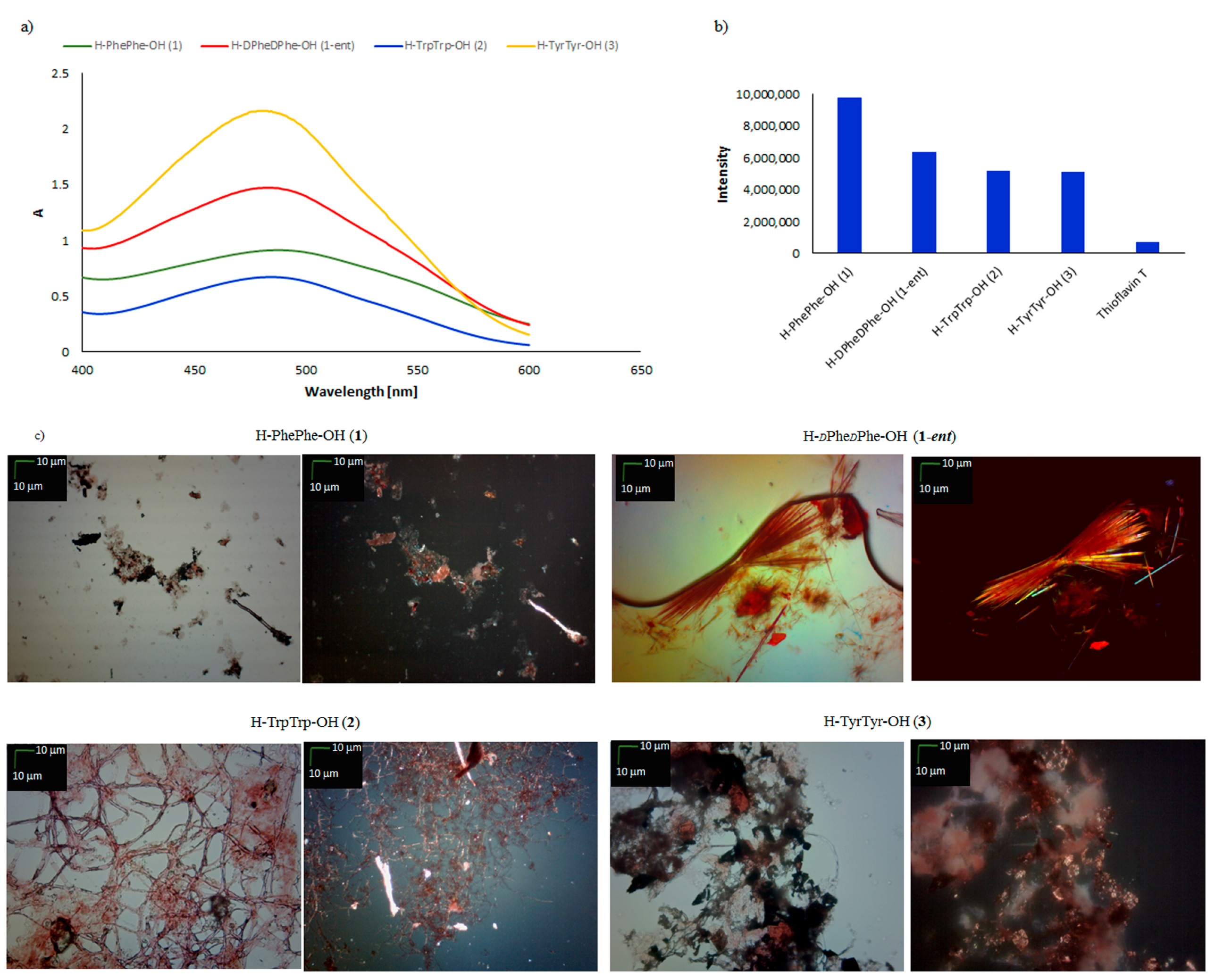
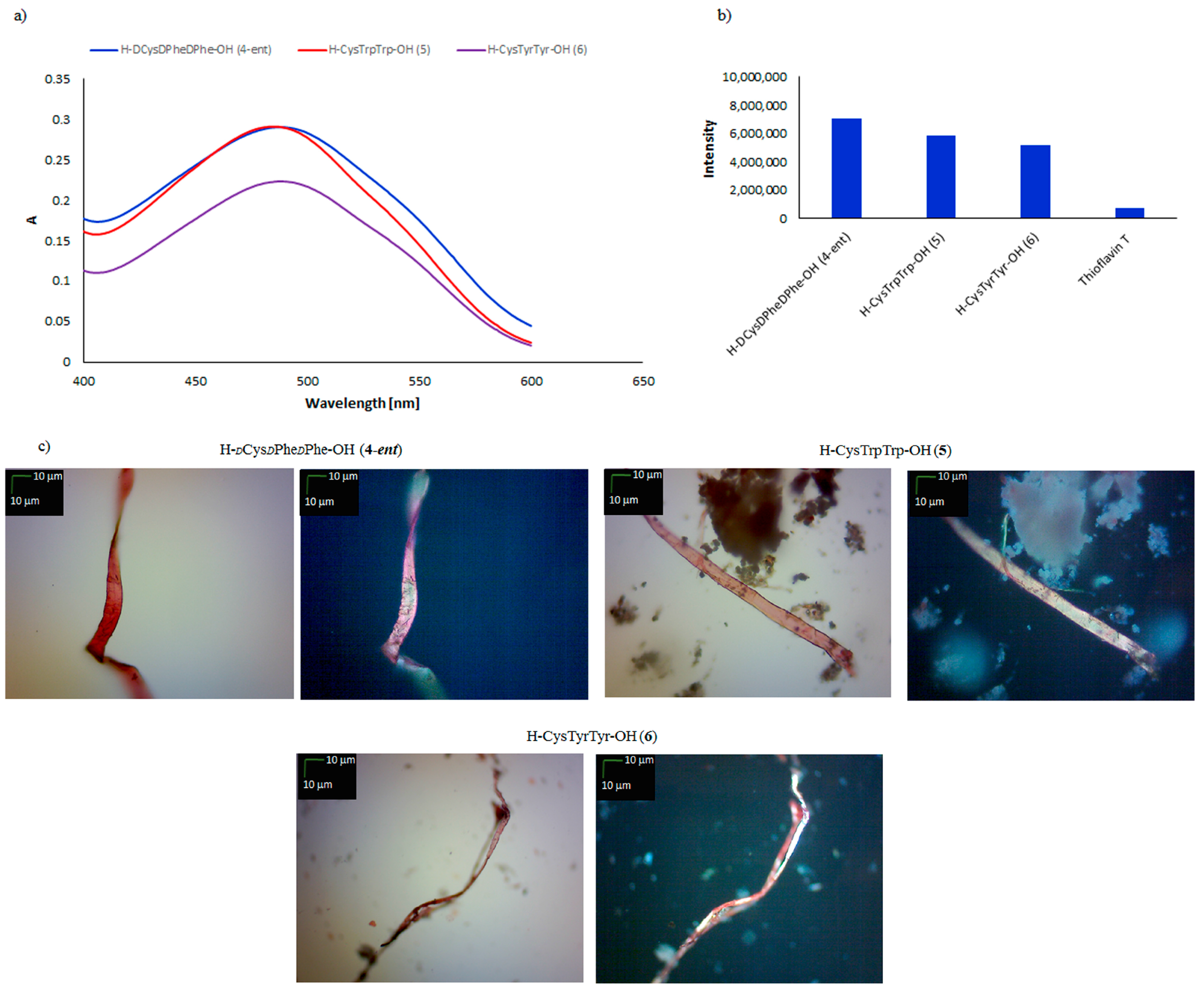
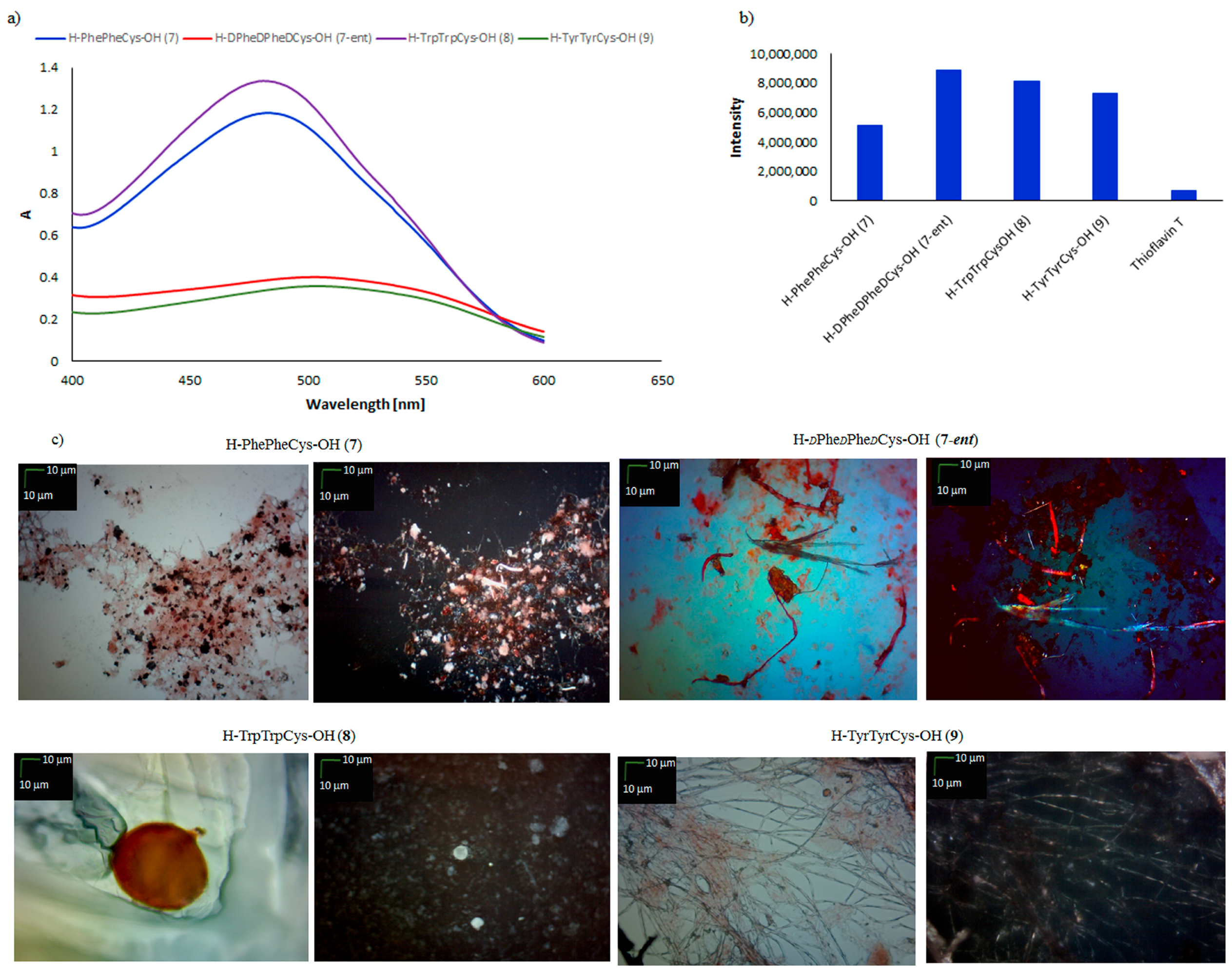
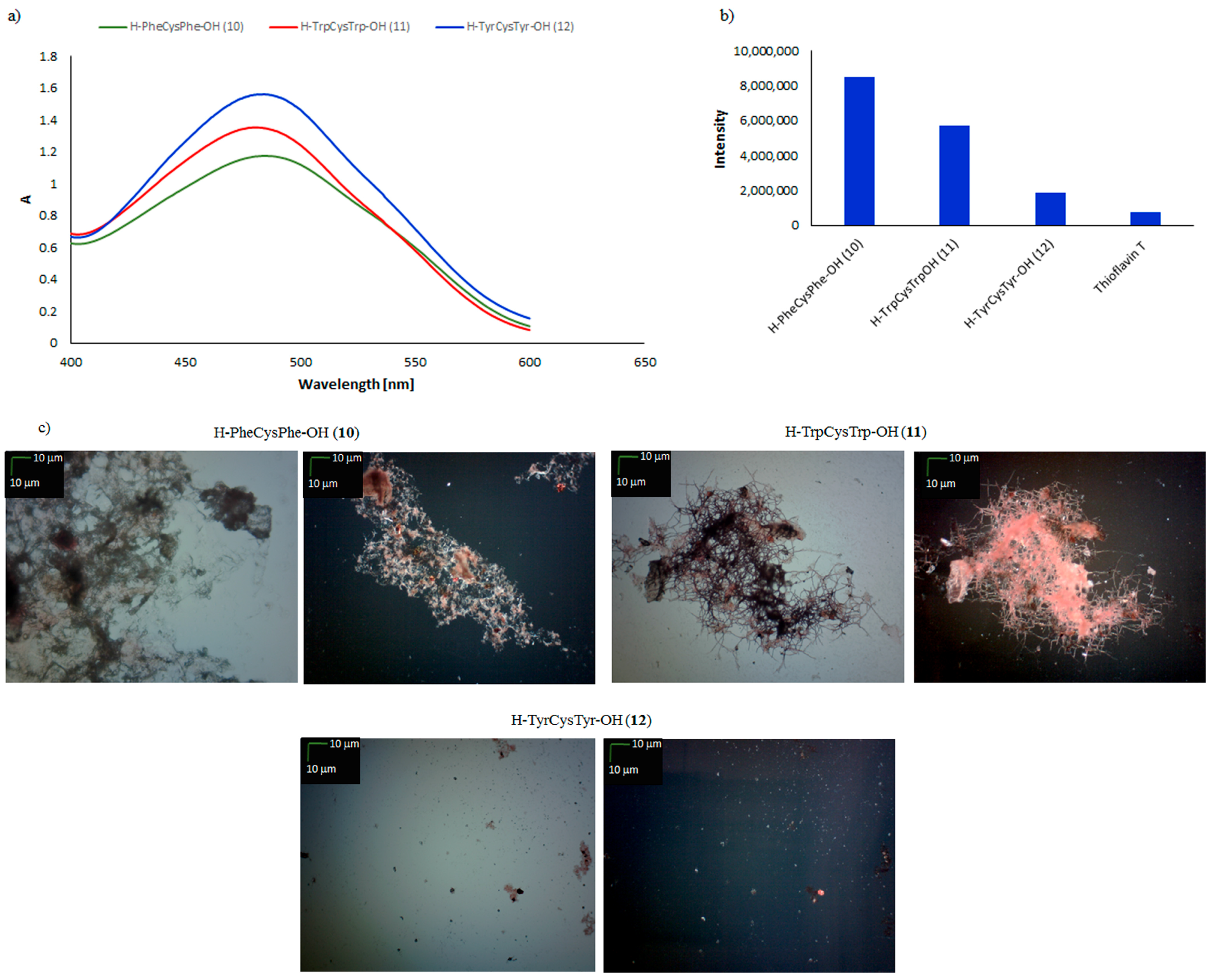
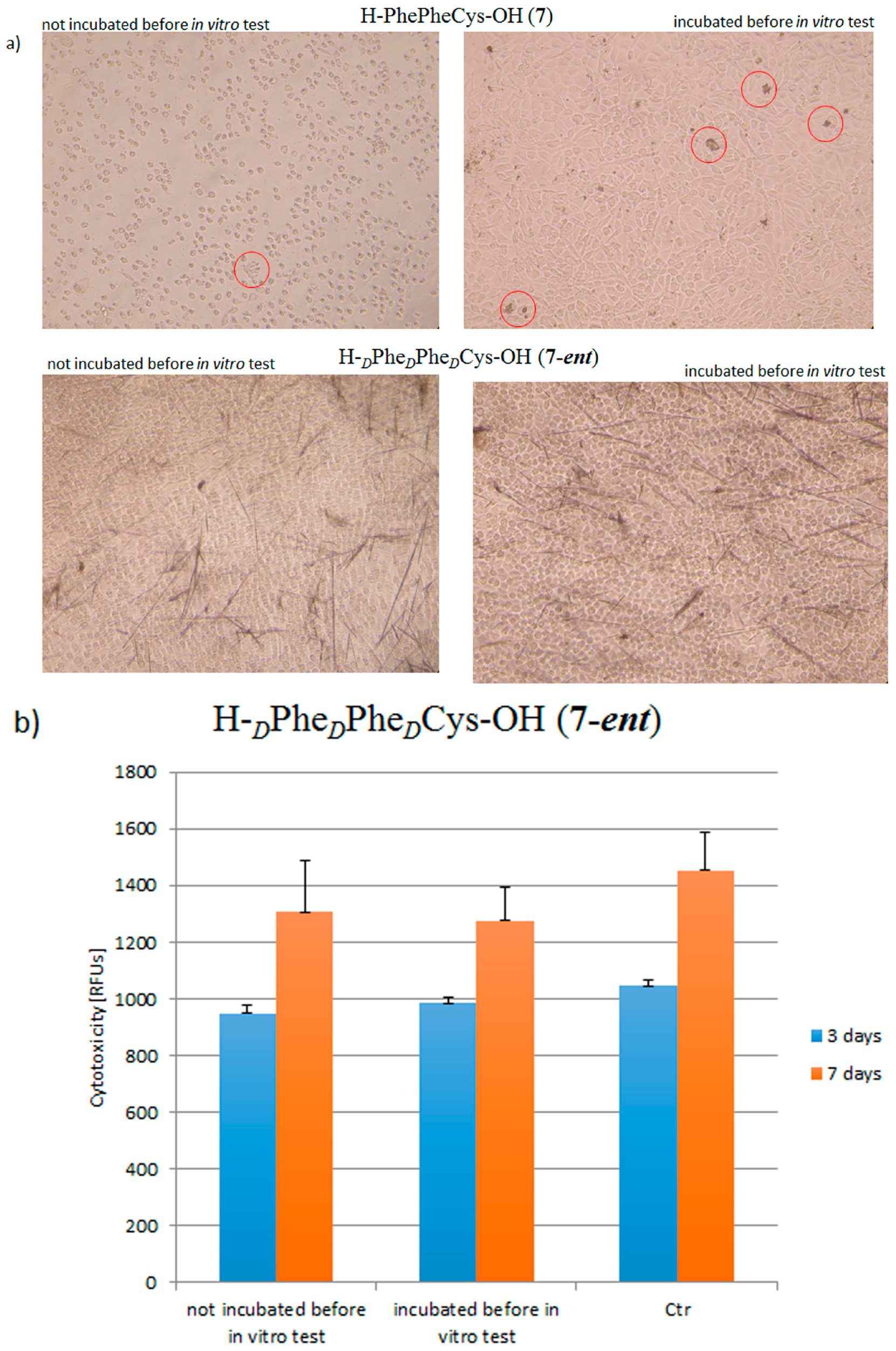
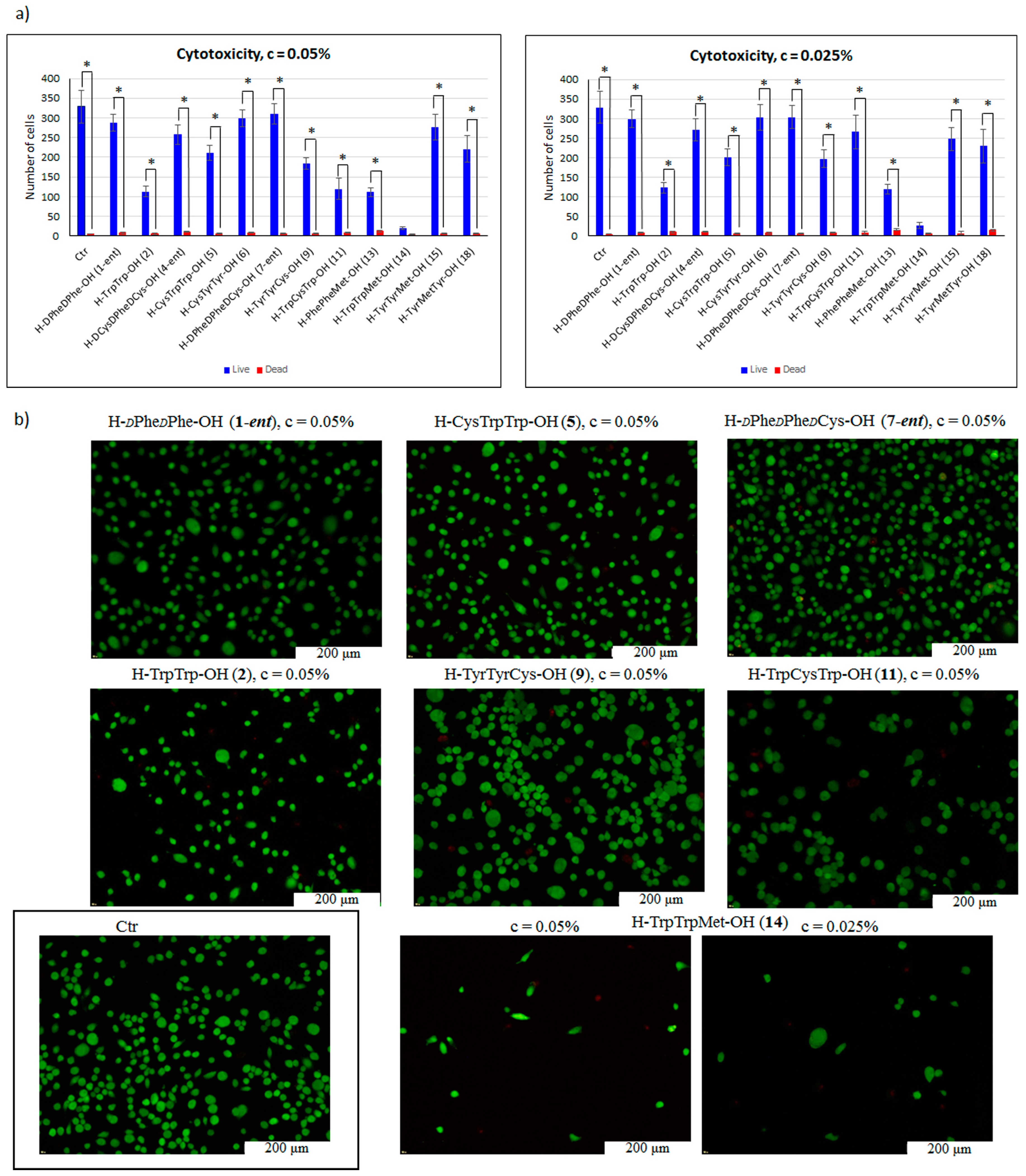
| Peptide | CR Assay | ThT Assay | Microscopic Examination, Morphology |
|---|---|---|---|
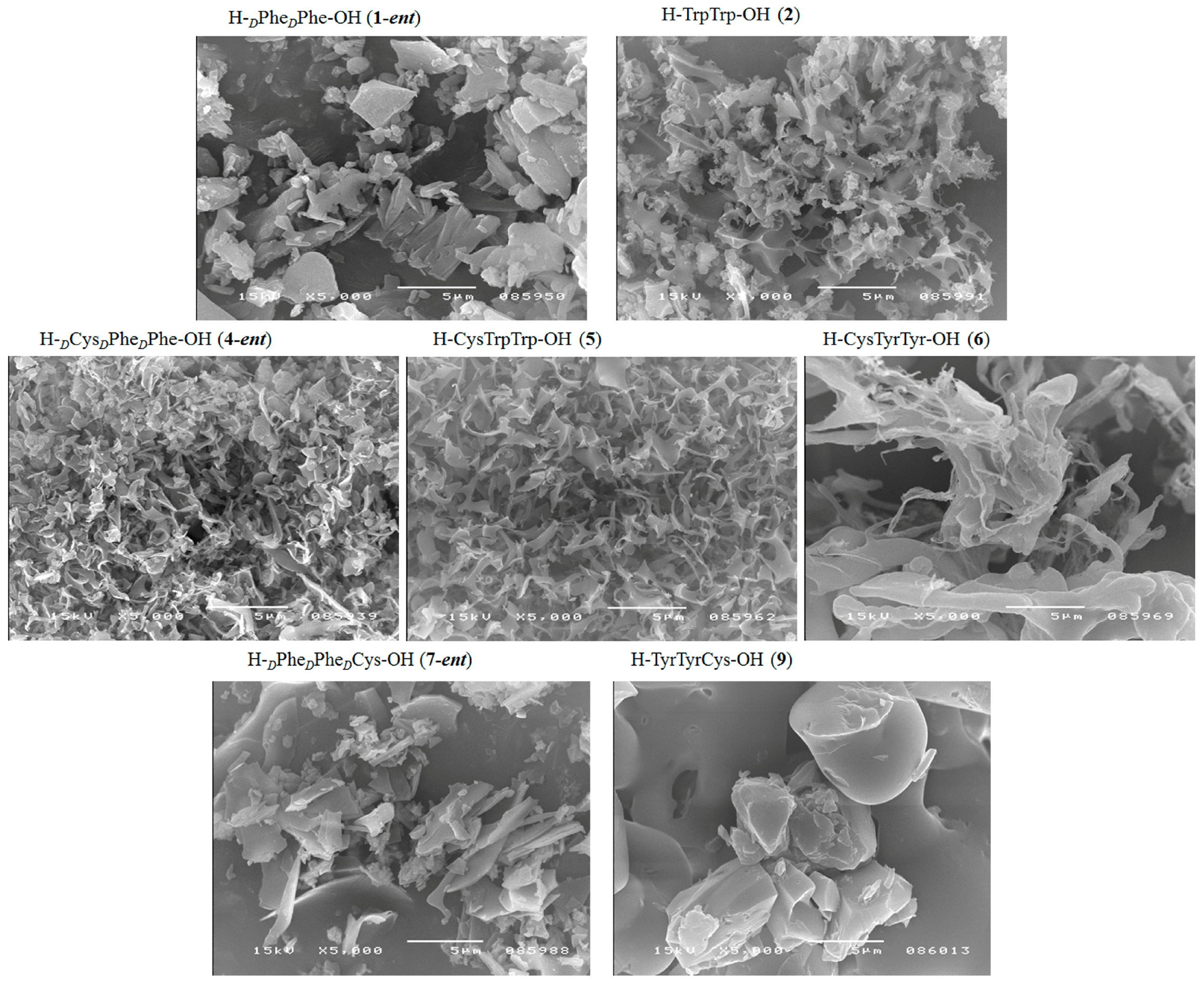
| H–PhePhe–OH (1) | + | + | + fibrous structure |
| H–dPhedPhe–OH (1-ent) | + | + | + fibrous structure |
| H–TrpTrp–OH (2) | + | + | + fibrous structure |
| H–TyrTyr–OH (3) | +/− | + | +/− cluster of fibrous and amorphous structures |
| H–dCysdPhedPhe–OH (4-ent) | + | + | + fibrous structure |
| H–CysTrpTrp–OH (5) | + | + | + fibrous structure |
| H–CysTyrTyr–OH (6) | + | + | + fibrous structure |
| H–PhePheCys–OH (7) | +/− | +/− | +/− cluster of fibrous and amorphous structures |
| H–dPhedPhedCys–OH (7-ent) | + | + | + fibrous structure |
| H–TrpTrpCys–OH (8) | +/− | + | +/− sphere |
| H–TyrTyrCy–OH (9) | + | + | + fibrous structure |
| H–PheCysPhe–OH (10) | + | + | +/− cluster of fibrous and amorphous structures |
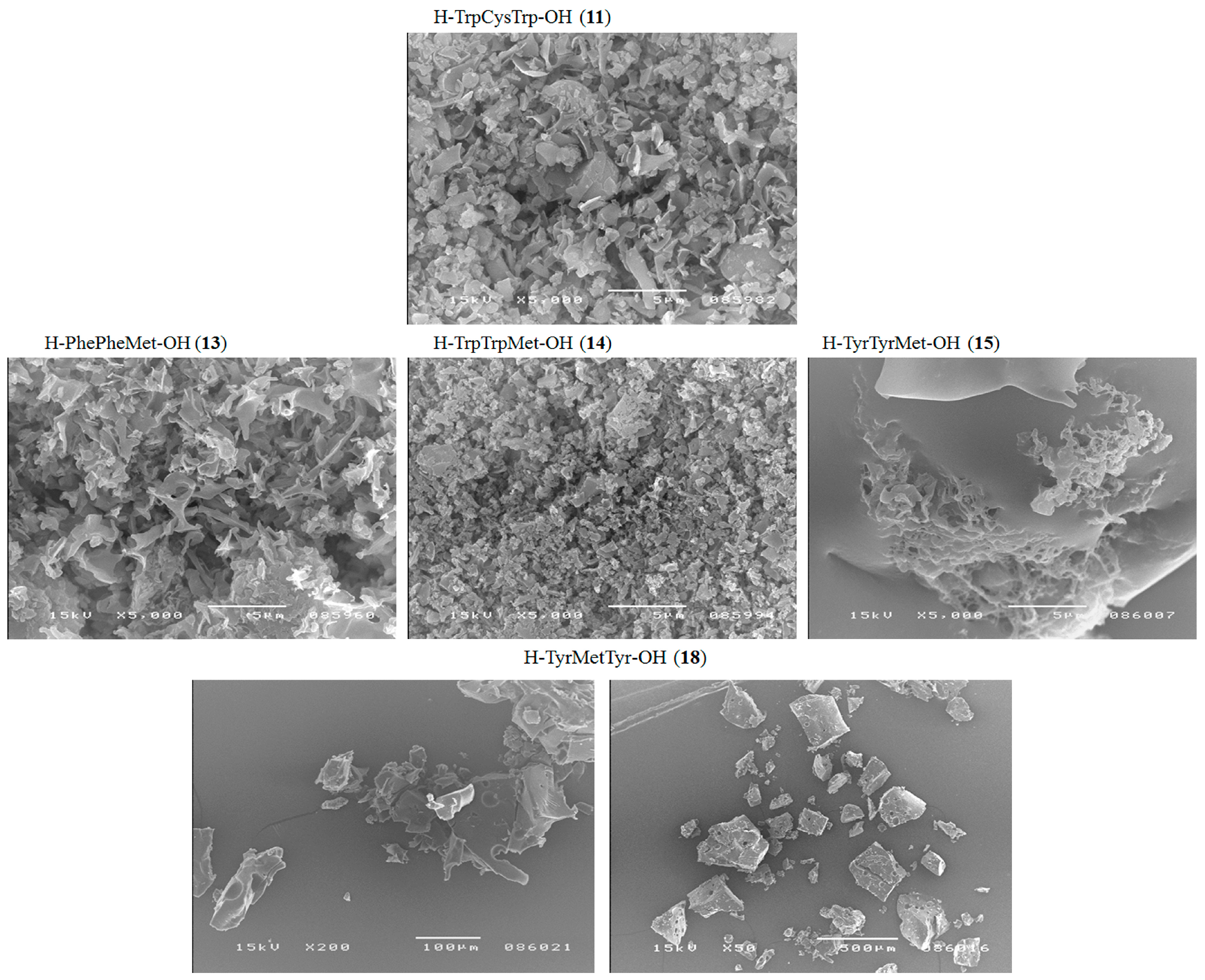
| H–TrpCysTrp–OH (11) | + | + | +/− fibrous structure |
| H–TyrCysTyr–OH (12) | − | − | − |
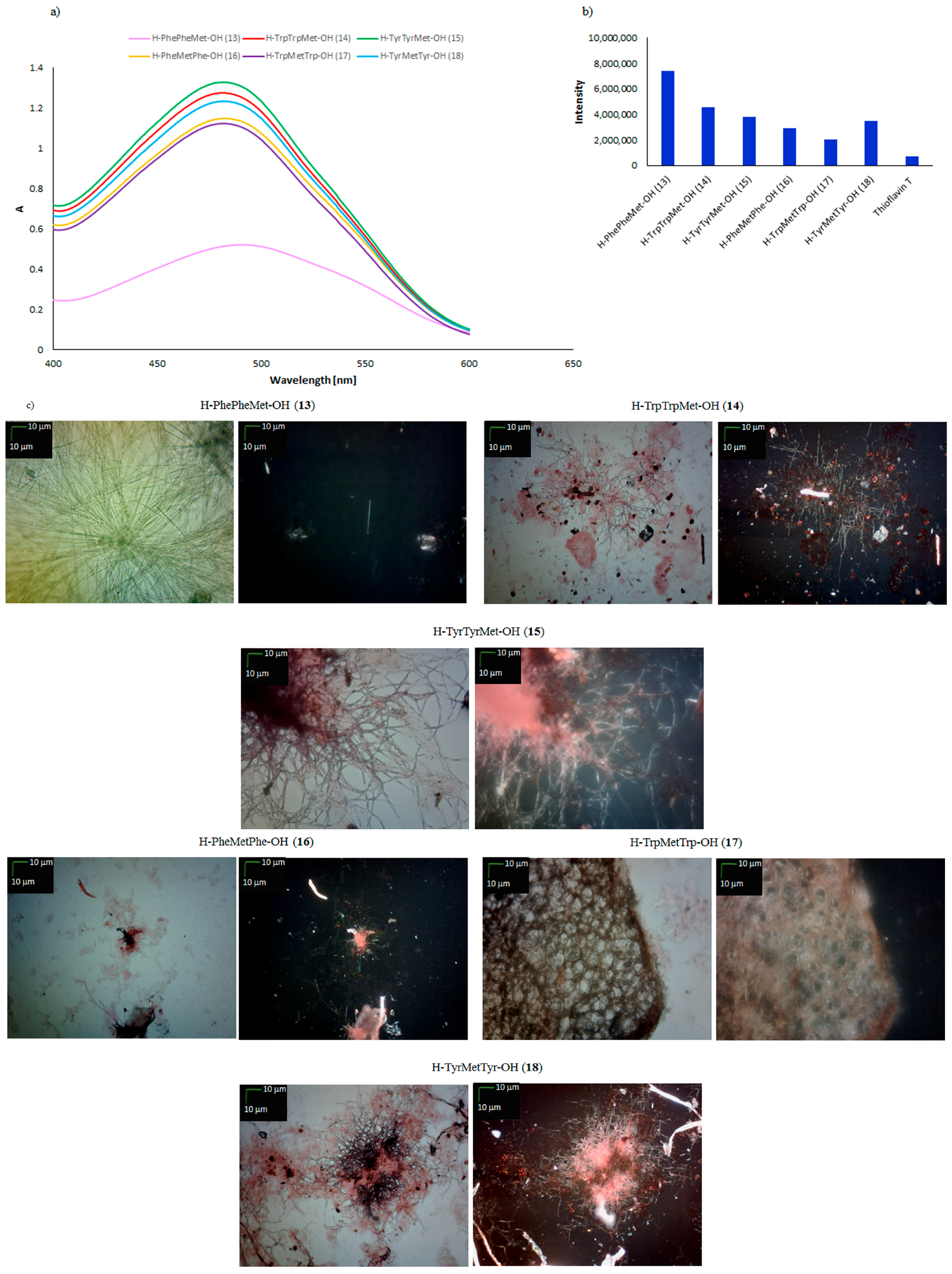
| H–PhePheMet–OH (13) | + | + | + fibrous structure |
| H–TrpTrpMet–OH (14) | +/− | +/− | +/− cluster of fibrous and amorphous structures |
| H–TyrTyrMet–OH (15) | +/− | +/− | +/− cluster of fibrous and amorphous structures |
| H–PheMetPhe–OH (16) | +/− | +/− | +/− cluster of fibrous and amorphous structures |
| H–TrpMetTrp–OH (17) | +/− | − | − amorphous structures |
| H–TyrMetTyr–OH (18) | +/− | +/− | +/− cluster of fibrous and amorphous structures |
| Peptide | THETA (°) 1 |
|---|---|
| H–dPhedPhe–OH (1-ent) | 34.1 |
| H–TrpTrp–OH (2) | 42.1 |
| H–dCysdPhedPhe–OH (4-ent) | 39.2 |
| H–CysTrpTrp–OH (5) | 46.8 |
| H–CysTyrTyr–OH (6) | 14.3 |
| H–dPhedPhedCys–OH (7-ent) | 40.1 |
| H–TyrTyrCys–OH (9) | 11.2 |
| H–TrpCysTrp–OH (11) | 49.3 |
| H–PhePheMet–OH (13) | 27.5 |
| H–TrpTrpMet–OH (14) | 20.2 |
| H–TyrTyrMet–OH (15) | 11.6 |
| H–TyrMetTyr–OH (18) | 12.7 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fraczyk, J.; Lipinski, W.; Chaberska, A.; Wasko, J.; Rozniakowski, K.; Kaminski, Z.J.; Bogun, M.; Draczynski, Z.; Menaszek, E.; Stodolak-Zych, E.; et al. Search for Fibrous Aggregates Potentially Useful in Regenerative Medicine Formed under Physiological Conditions by Self-Assembling Short Peptides Containing Two Identical Aromatic Amino Acid Residues. Molecules 2018, 23, 568. https://doi.org/10.3390/molecules23030568
Fraczyk J, Lipinski W, Chaberska A, Wasko J, Rozniakowski K, Kaminski ZJ, Bogun M, Draczynski Z, Menaszek E, Stodolak-Zych E, et al. Search for Fibrous Aggregates Potentially Useful in Regenerative Medicine Formed under Physiological Conditions by Self-Assembling Short Peptides Containing Two Identical Aromatic Amino Acid Residues. Molecules. 2018; 23(3):568. https://doi.org/10.3390/molecules23030568
Chicago/Turabian StyleFraczyk, Justyna, Wojciech Lipinski, Agata Chaberska, Joanna Wasko, Kamil Rozniakowski, Zbigniew J. Kaminski, Maciej Bogun, Zbigniew Draczynski, Elzbieta Menaszek, Ewa Stodolak-Zych, and et al. 2018. "Search for Fibrous Aggregates Potentially Useful in Regenerative Medicine Formed under Physiological Conditions by Self-Assembling Short Peptides Containing Two Identical Aromatic Amino Acid Residues" Molecules 23, no. 3: 568. https://doi.org/10.3390/molecules23030568





