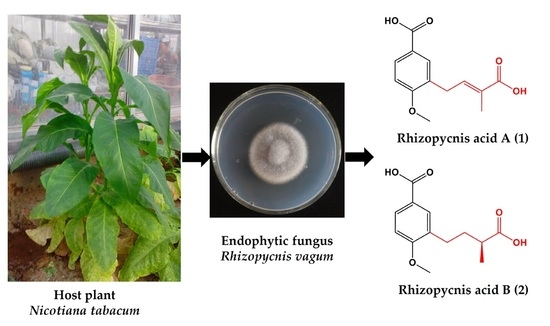
Two New Anisic Acid Derivatives from Endophytic Fungus Rhizopycnis vagum Nitaf22 and Their Antibacterial Activity
Abstract
Share and Cite
Wang, A.; Li, P.; Zhang, X.; Han, P.; Lai, D.; Zhou, L. Two New Anisic Acid Derivatives from Endophytic Fungus Rhizopycnis vagum Nitaf22 and Their Antibacterial Activity. Molecules 2018, 23, 591. https://doi.org/10.3390/molecules23030591
Wang A, Li P, Zhang X, Han P, Lai D, Zhou L. Two New Anisic Acid Derivatives from Endophytic Fungus Rhizopycnis vagum Nitaf22 and Their Antibacterial Activity. Molecules. 2018; 23(3):591. https://doi.org/10.3390/molecules23030591
Chicago/Turabian StyleWang, Ali, Peng Li, Xuping Zhang, Peipei Han, Daowan Lai, and Ligang Zhou. 2018. "Two New Anisic Acid Derivatives from Endophytic Fungus Rhizopycnis vagum Nitaf22 and Their Antibacterial Activity" Molecules 23, no. 3: 591. https://doi.org/10.3390/molecules23030591
APA StyleWang, A., Li, P., Zhang, X., Han, P., Lai, D., & Zhou, L. (2018). Two New Anisic Acid Derivatives from Endophytic Fungus Rhizopycnis vagum Nitaf22 and Their Antibacterial Activity. Molecules, 23(3), 591. https://doi.org/10.3390/molecules23030591