Atypical Kinetics and Albumin Effect of Glucuronidation of 5-n-Butyl-4-{4-[2-(1H-tetrazole-5- yl)-1H-pyrrol-1-yl]phenylmethyl}-2,4-dihydro-2-(2,6- dichlorophenyl)-3H-1,2,4-triazol-3-one, a Novel Nonpeptide Angiotensin Type 1 Receptor Antagonist, in Liver Microsomes and UDP-Glucuronosyl-transferase
Abstract
:1. Introduction
2. Results
2.1. Metabolites of Ib in Microsomes
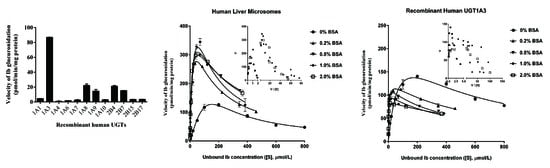
2.2. Screening by Recombinant UGTs
2.3. Binding of Ib to BSA
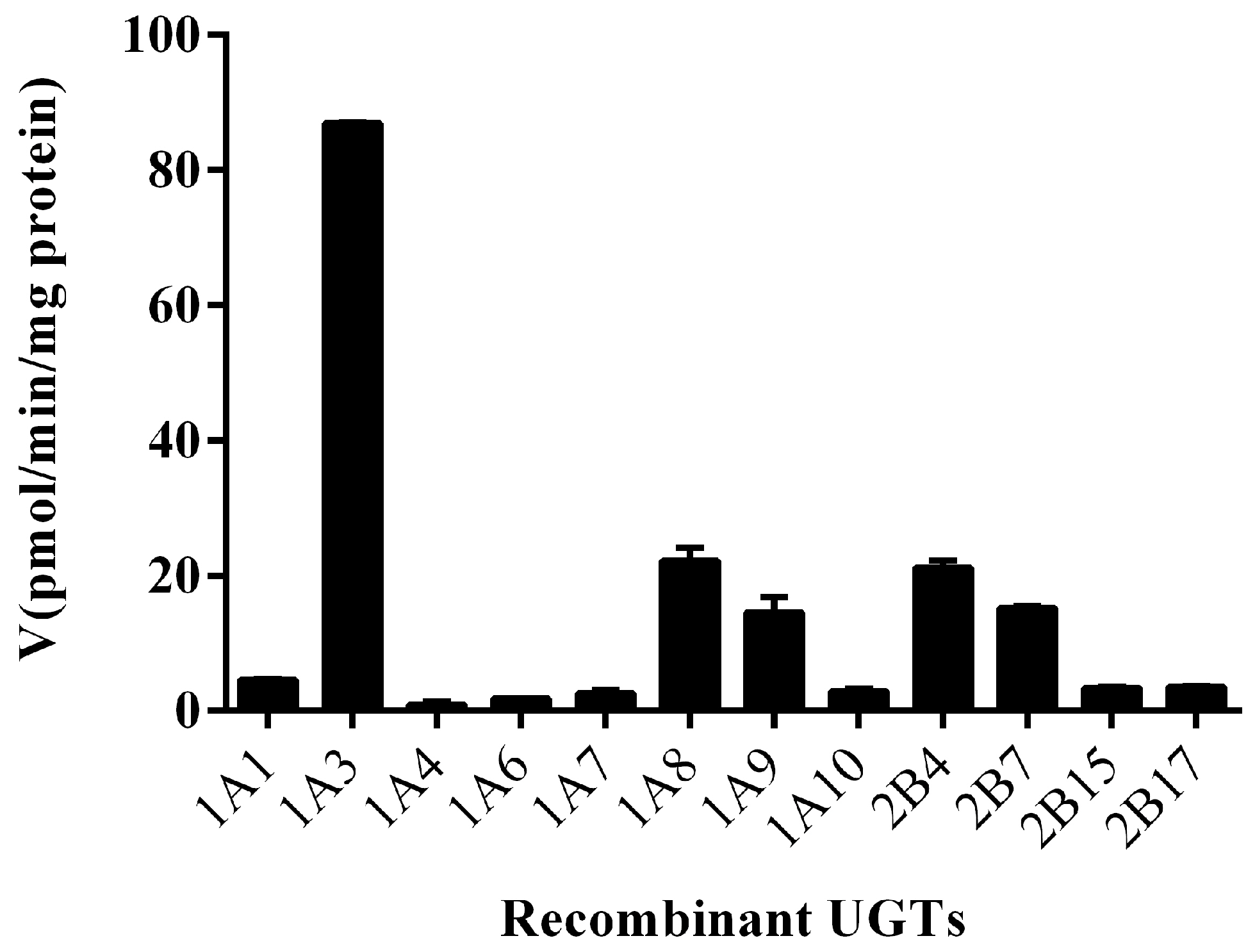
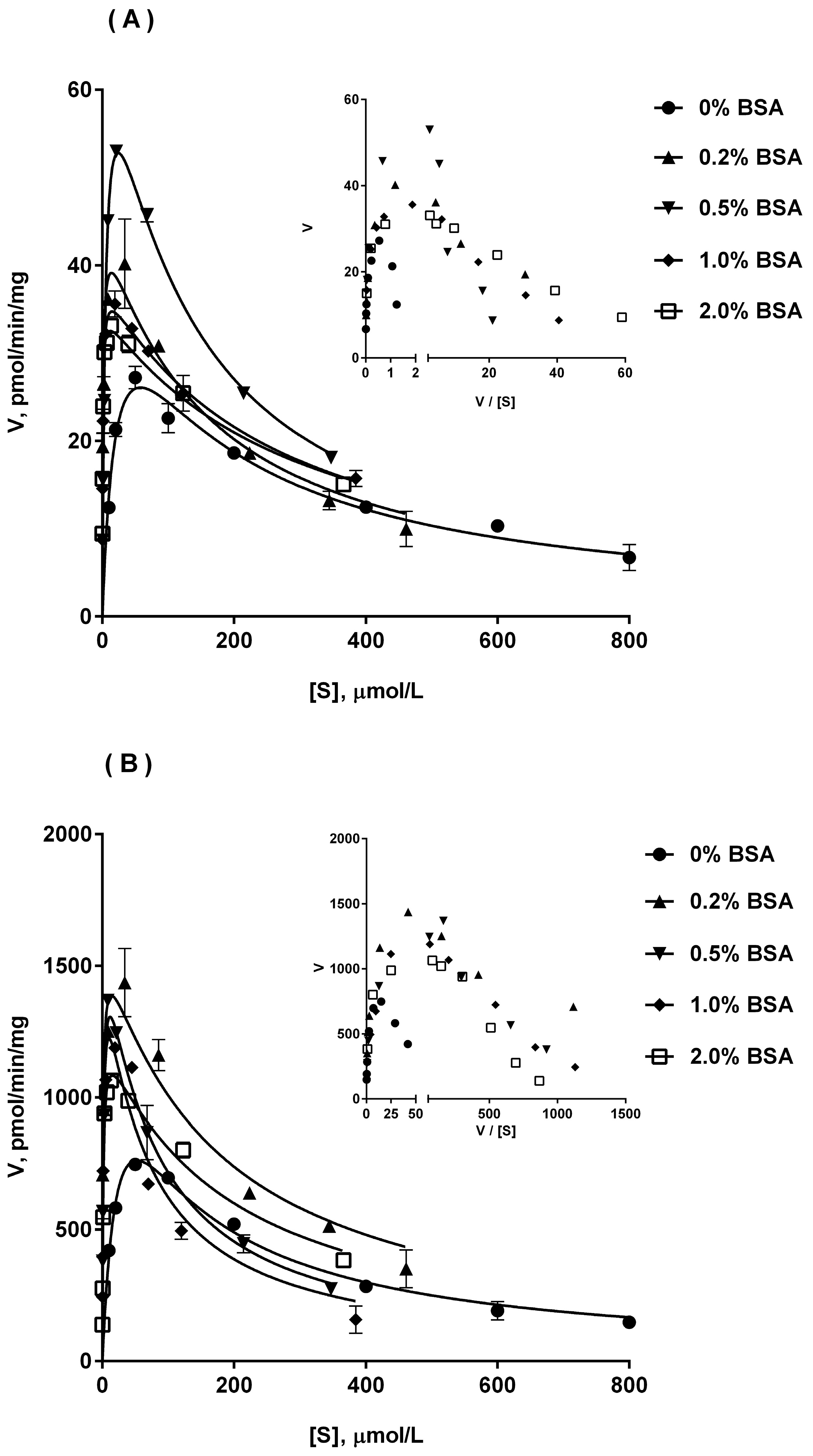
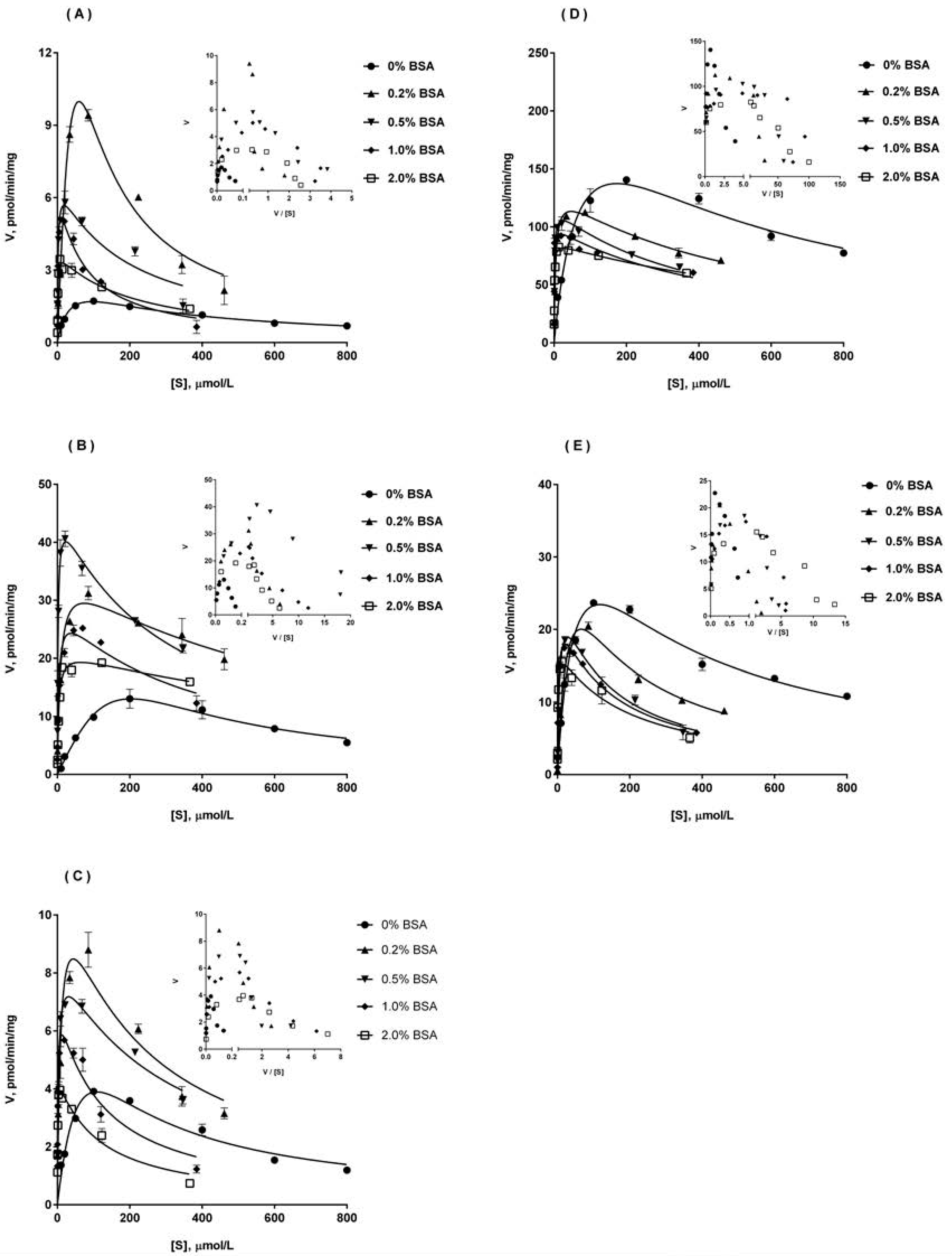
2.4. Ib Glucuronidation by Microsomes and BSA Effect
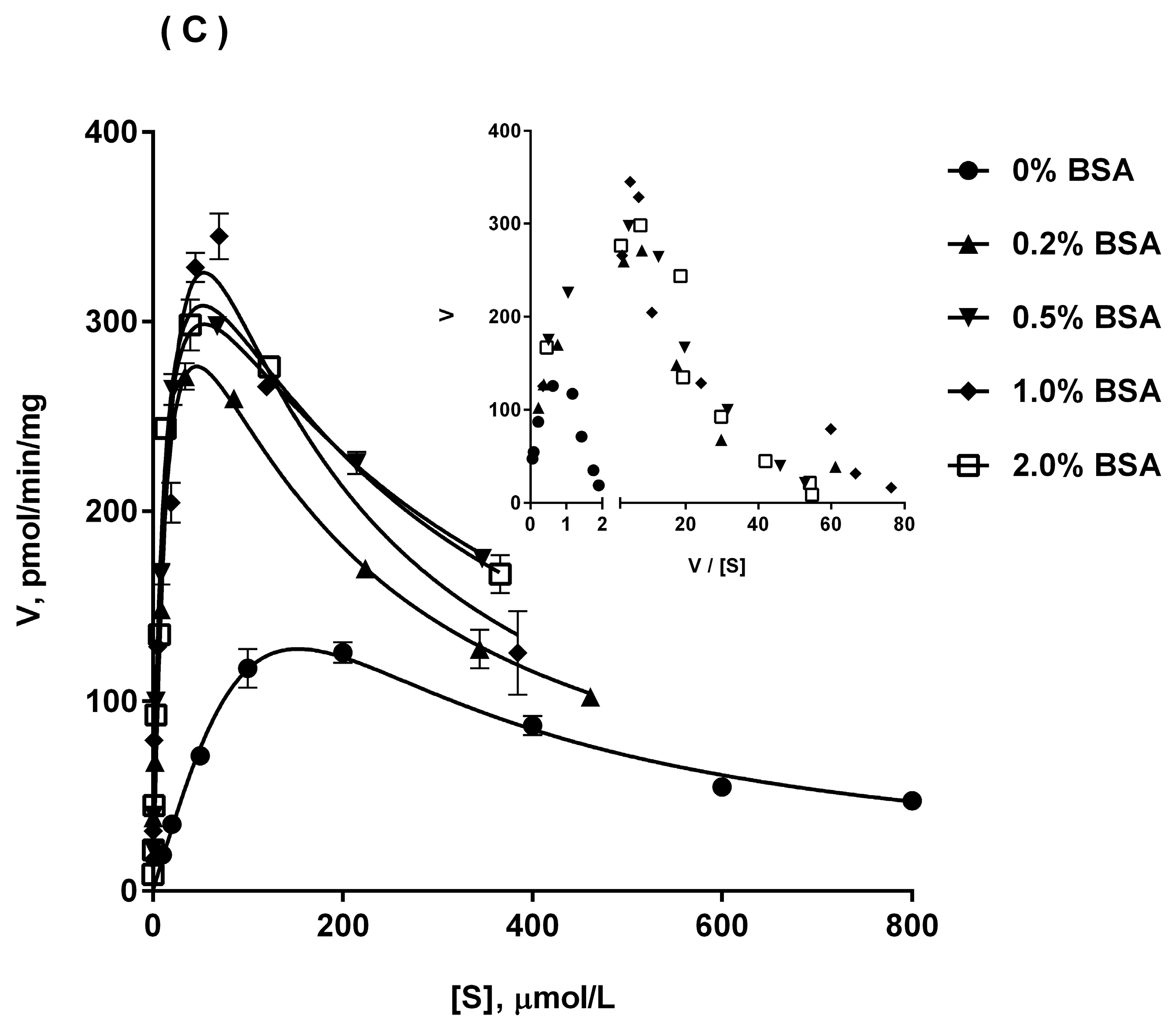
2.5. Ib Glucuronidation by Recombinant UGTs and BSA Effect
3. Discussion
4. Materials and Methods
4.1. Chemicals and Materials
4.2. LC-MS/MS Assay
4.3. Enzyme Incubation
4.4. Glucuronidation in Microsomes
4.5. Glucuronidation by Recombinant UGTs
4.6. Protein Binding Assay of Ib
4.7. Enzyme Kinetic Data Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| Ib | 5-n-butyl-4-{4-[2-(1H-tetrazole-5-yl)-1H-pyrrol-1-yl]phenylmethyl}-2,4-dihydro-2-(2,6-dichloro- phenyl)-3H-1,2,4-triazol-3-one |
| AT1 | angiotensin type 1 |
| BSA | bovine serum albumin |
| IS | internal standard |
| UPDGA | uridine 5’-diphosphoglucuronic acid |
| NADP | β-nicotinamide adenine dinucleotide phosphate |
| G-6-P | d-glucose 6-phosphate |
| PDH | glucose-6-phosphate dehydrogenase |
| HLM | human liver microsomes |
| MKLM | monkey liver microsomes |
| DLM | dog liver microsomes |
| RLM | rat liver microsomes |
| MLM | mouse liver microsomes |
| UGT | UDP-glucuronosyltransferases |
| CYP | cytochrome P450 |
| LC-MS/MS | liquid chromatography-tandem mass spectrometry |
| DDI | drug-drug interactions |
| IVIVE | in vitro-in vivo extrapolation |
References
- Xu, J.Y.; Zhou, Y.X.; Wu, X.M.; Tang, K.; Wang, Q.J. Synthesis and bioactivity of 1-aryl-3-alkyl-1,4-dihydro-4-substitued-5h-1,2,4,-triazolinones-5. J. China Pharm. Univ. 1999, 30, 323–327. [Google Scholar]
- Wu, J.H.; Wang, Q.J.; Hu, S.; Xu, J.Y.; Wu, X.M. Antihypertensive effects of a new angiotensin type 1 receptor antagonist ib on rats. J. China Pharm. Univ. 2005, 36, 563–566. [Google Scholar]
- Zhang, J.; Wang, Q.J.; Xu, J.Y.; Wu, X.M. Effects of a new angiotensin type 1 receptor antagonist Ib on the cardiac hypertrophy induced by l-thyroxin in rats. J. China Pharm. Univ. 2006, 37, 157–160. [Google Scholar]
- Wang, Y.; Wang, W.; Wang, Q.; Wu, J.; Xu, J.; Wu, X. [Ca2+]i and PKC-alpha are involved in the inhibitory effects of Ib, a novel nonpeptide angiotensinii subtype AT1 receptor antagonist, on angiotensinii-induced vascular contraction in vitro. Biochem. Biophys. Res. Commun. 2007, 364, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yan, T.; Wang, Q.; Wang, W.; Xu, J.; Wu, X.; Ji, H. Pkc-dependent extracellular signal-regulated kinase 1/2 pathway is involved in the inhibition of Ib on angiotensinii-induced proliferation of vascular smooth muscle cells. Biochem. Biophys. Res. Commun. 2008, 375, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Wang, G.; Sun, J.; Li, X.; Zheng, Y.; Ai, H.; Lv, H.; Wu, X.; Xu, J. Identification of the major metabolites of 5-n-butyl-4-{4-[2-(1H-tetrazole-5-yl)-1H-pyrrol-1-yl]phenylmethyl}-2,4-dihydro-2- (2,6-dichloridephenyl)-3H-1,2,4-triazol-3-one, a new angiotensin type 1 receptor antagonist, in rat bile by HPLC-diode array detection-MS and HPLC-MS/MS. Biomed. Chromatogr. 2007, 21, 912–924. [Google Scholar] [PubMed]
- Tang, C.; Lin, Y.; Rodrigues, A.D.; Lin, J.H. Effect of albumin on phenytoin and tolbutamide metabolism in human liver microsomes: An impact more than protein binding. Drug Metab. Dispos. 2002, 30, 648–654. [Google Scholar] [CrossRef] [PubMed]
- Rowland, A.; Elliot, D.J.; Knights, K.M.; Mackenzie, P.I.; Miners, J.O. The “albumin effect” and in vitro-in vivo extrapolation: Sequestration of long-chain unsaturated fatty acids enhances phenytoin hydroxylation by human liver microsomal and recombinant cytochrome P450 2C9. Drug Metab. Dispos. 2008, 36, 870–877. [Google Scholar] [CrossRef] [PubMed]
- Wattanachai, N.; Polasek, T.M.; Heath, T.M.; Uchaipichat, V.; Tassaneeyakul, W.; Tassaneeyakul, W.; Miners, J.O. In vitro–in vivo extrapolation of CYP2C8-catalyzed paclitaxel 6α-hydroxylation: Effects of albumin on in vitro kinetic parameters and assessment of interindividual variability in predicted clearance. Eur. J. Clin. Pharmacol. 2011, 67, 815–824. [Google Scholar] [CrossRef] [PubMed]
- Wattanachai, N.; Tassaneeyakul, W.; Rowland, A.; Elliot, D.J.; Bowalgaha, K.; Knights, K.M.; Miners, J.O. Effect of albumin on human liver microsomal and recombinant CYP1A2 activities: Impact on in vitro-in vivo extrapolation of drug clearance. Drug Metab. Dispos. 2012, 40, 982–989. [Google Scholar] [CrossRef] [PubMed]
- Rowland, A.; Gaganis, P.; Elliot, D.J.; Mackenzie, P.I.; Knights, K.M.; Miners, J.O. Binding of inhibitory fatty acids is responsible for the enhancement of UDP-glucuronosyltransferase 2B7 activity by albumin: Implications for in vitro-in vivo extrapolation. J. Pharmacol. Exp. Ther. 2007, 321, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Rowland, A.; Knights, K.M.; Mackenzie, P.I.; Miners, J.O. The “albumin effect” and drug glucuronidation: Bovine serum albumin and fatty acid-free human serum albumin enhance the glucuronidation of UDP-glucuronosyltransferase (UGT) 1A9 substrates but not UGT1A1 and UGT1A6 activities. Drug Metab. Dispos. 2008, 36, 1056–1062. [Google Scholar] [CrossRef] [PubMed]
- Raungrut, P.; Uchaipichat, V.; Elliot, D.J.; Janchawee, B.; Somogyi, A.A.; Miners, J.O. In vitro-in vivo extrapolation predicts drug-drug interactions arising from inhibition of codeine glucuronidation by dextropropoxyphene, fluconazole, ketoconazole, and methadone in humans. J. Pharmacol. Exp. Ther. 2010, 334, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Manevski, N.; Moreolo, P.S.; Yli-Kauhaluoma, J.; Finel, M. Bovine serum albumin decreases km values of human UDP-glucuronosyltransferases 1A9 and 2B7 and increases vmax values of UGT1A9. Drug Metab. Dispos. 2011, 39, 2117–2129. [Google Scholar] [CrossRef] [PubMed]
- Stearns, R.A.; Doss, G.A.; Miller, R.R.; Chiu, S.H. Synthesis and identification of a novel tetrazole metabolite of the angiotensin ii receptor antagonist DuP 753. Drug Metab. Dispos. 1991, 19, 1160–1162. [Google Scholar] [PubMed]
- Izukawa, T.; Nakajima, M.; Fujiwara, R.; Yamanaka, H.; Fukami, T.; Takamiya, M.; Aoki, Y.; Ikushiro, S.; Sakaki, T.; Yokoi, T. Quantitative analysis of UDP-glucuronosyltransferase (UGT) 1A and UGT2B expression levels in human livers. Drug Metab. Dispos. 2009, 37, 1759–1768. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S.; Nakajin, S. Determination of mrna expression of human udp-glucuronosyltransferases and application for localization in various human tissues by real-time reverse transcriptase-polymerase chain reaction. Drug Metab. Dispos. 2009, 37, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Bowalgaha, K.; Elliot, D.J.; Mackenzie, P.I.; Knights, K.M.; Swedmark, S.; Miners, J.O. S-naproxen and desmethylnaproxen glucuronidation by human liver microsomes and recombinant human UDP-glucuronosyltransferases (UGT): Role of UGT2B7 in the elimination of naproxen. Br. J. Clin. Pharmacol. 2005, 60, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Court, M.H.; Krishnaswamy, S.; Hao, Q.; Duan, S.X.; Patten, C.J.; Von Moltke, L.L.; Greenblatt, D.J. Evaluation of 3'-azido-3'-deoxythymidine, morphine, and codeine as probe substrates for UDP-glucuronosyltransferase 2B7 (UGT2B7) in human liver microsomes: Specificity and influence of the UGT2B7*2 polymorphism. Drug Metab. Dispos. 2003, 31, 1125–1133. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Sun, J.; Peng, Y.; Zhang, R.; Shao, F.; Hu, X.; Zhu, J.; Wang, X.; Cheng, X.; Zhu, Y.; et al. Glucuronidation of edaravone by human liver and kidney microsomes: Biphasic kinetics and identification of UGT1A9 as the major udp-glucuronosyltransferase isoform. Drug Metab. Dispos. 2012, 40, 734–741. [Google Scholar] [CrossRef] [PubMed]
- Tsoutsikos, P.; Miners, J.O.; Stapleton, A.; Thomas, A.; Sallustio, B.C.; Knights, K.M. Evidence that unsaturated fatty acids are potent inhibitors of renal UDP-glucuronosyltransferases (UGT): Kinetic studies using human kidney cortical microsomes and recombinant UGT1A9 and UGT2B7. Biochem. Pharmacol. 2004, 67, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Houston, J.B. Utility of in vitro drug metabolism data in predicting in vivo metabolic clearance. Biochem. Pharmacol. 1994, 47, 1469–1479. [Google Scholar] [CrossRef]
- Moltke, L.L.V.; Greenblatt, D.J.; Schmider, J.; Wright, C.E.; Harmatz, J.S.; Shader, R.I. In vitro approaches to predicting drug interactions in vivo. Biochem. Pharmacol. 1998, 55, 113–122. [Google Scholar] [CrossRef]
- Miners, J.O.; Mackenzie, P.I.; Knights, K.M. The prediction of drug-glucuronidation parameters in humans: UDP-glucuronosyltransferase enzyme-selective substrate and inhibitor probes for reaction phenotyping and in vitro-in vivo extrapolation of drug clearance and drug-drug interaction potential. Drug Metab. Rev. 2010, 42, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Hosea, N.A.; Collard, W.T.; Bsc, S.C.; PharmD, T.S.M.; Fang, R.X.; Jones, H.; Kakar, S.M.; Nakai, Y.; Smith, B.J.; Bsc, R.W. Prediction of human pharmacokinetics from preclinical information: Comparative accuracy of quantitative prediction approaches. J. Clin. Pharmacol. 2009, 49, 513–533. [Google Scholar] [CrossRef] [PubMed]
- Ludden, L.K.; Ludden, T.M.; Collins, J.M.; Pentikis, H.S.; Strong, J.M. Effect of albumin on the estimation, in vitro, of phenytoin vmax and km values: Implications for clinical correlation. J. Pharmacol. Exp. Ther. 1997, 282, 391–396. [Google Scholar] [PubMed]
- Shou, M.; Lin, Y.; Lu, P.; Tang, C.; Mei, Q.; Cui, D.; Tang, W.; Ngui, J.S.; Lin, C.C.; Singh, R. Enzyme kinetics of cytochrome p450-mediated reactions. Curr. Drug Metab. 2001, 2, 17. [Google Scholar] [CrossRef] [PubMed]
- Hutzler, J.M.; Tracy, T.S. Atypical kinetic profiles in drug metabolism reactions. Drug Metab. Dispos. 2002, 30, 355–362. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds of Ib and Ib monoglucuronide are available from the authors. |

| Ib, μM | 0.2% BSA | 0.5% BSA | 1% BSA | 2% BSA |
|---|---|---|---|---|
| 10 | 0.063 ± 0.005 | 0.041 ± 0.003 | 0.022 ± 0.002 | 0.016 ± 0.001 |
| 20 | 0.114 ± 0.010 | 0.043 ± 0.004 | 0.024 ± 0.002 | 0.020 ± 0.001 |
| 50 | 0.169 ± 0.013 | 0.063 ± 0.005 | 0.026 ± 0.002 | 0.021 ± 0.002 |
| 100 | 0.339 ± 0.033 | 0.084 ± 0.009 | 0.053 ± 0.004 | 0.031 ± 0.002 |
| 200 | 0.428 ± 0.047 | 0.105 ± 0.010 | 0.095 ± 0.007 | 0.035 ± 0.003 |
| 400 | 0.559 ± 0.038 | 0.170 ± 0.017 | 0.112 ± 0.009 | 0.043 ± 0.005 |
| 600 | 0.574 ± 0.051 | 0.358 ± 0.028 | 0.117 ± 0.010 | 0.066 ± 0.007 |
| 800 | 0.577 ± 0.040 | 0.434 ± 0.034 | 0.150 ± 0.012 | 0.153 ± 0.008 |
| 2000 | 0.613 ± 0.079 | 0.495 ± 0.039 | 0.192 ± 0.015 | 0.183 ± 0.010 |
| Enzyme Source | Parameter | No BSA | 0.2% BSA | 0.5% BSA | 1% BSA | 2% BSA |
|---|---|---|---|---|---|---|
| HLM | Km (μM) | 59.8 ± 4.6 | 5.55 ± 0.95 | 5.80 ± 1.80 | 6.10 ± 1.10 | 5.99 ± 0.93 |
| Vmax (pmol/min/mg) | 179.8 ± 6.7 | 316.3 ± 33.6 | 356.8 ± 37.2 | 389.5 ± 85.3 | 357.3 ± 34.4 | |
| Ki (μM) | 455.5 ± 59.9 | 300.3 ± 86.5 | 350.0 ± 106.6 | 200.3 ± 92.4 | 354.1 ± 106.5 | |
| CLint (μL/min/mg) | 3.02 ± 0.31 | 57.8 ± 9.6 | 64.4 ± 14.3 | 66.0 ± 20.6 | 61.2 ± 15.4 | |
| DLM | Km (μM) | 9.00 ± 2.10 | 1.48 ± 0.33 | 1.44 ± 0.20 | 1.40 ± 0.19 | 1.11 ± 0.23 |
| Vmax (pmol/min/mg) | 890.0 ± 44.0 | 1712.0 ± 308.1 | 1533.0 ± 122.81 | 1492.0 ± 71.4 | 1182.0 ± 65.4 | |
| Ki (μM) | 233.6 ± 54.0 | 199.0 ± 59.5 | 89.7 ± 19.9 | 74.1 ± 10.0 | 226.4 ± 20.0 | |
| CLint (μL/min/mg) | 101.9 ± 19.4 | 1164.5 ± 52.8 | 1086.6 ± 38.5 | 1083.7 ± 99.9 | 1088.2 ± 70.2 | |
| RLM | Km (μM) | 13.0 ± 3.1 | 1.83 ± 0.78 | 2.00 ± 0.12 | 1.92 ± 0.51 | 1.80 ± 0.40 |
| Vmax (pmol/min/mg) | 28.5 ± 2.4 | 41.5 ± 2.3 | 57.2 ± 10.3 | 37.5 ± 2.0 | 36.5 ± 3.7 | |
| Ki (μM) | 332.9 ± 50.3 | 187.5 ± 34.3 | 176.2 ± 84.6 | 270.0 ± 53.9 | 263.7 ± 58.0 | |
| CLint (μL/min/mg) | 2.25 ± 0.36 | 25.6 ± 5.6 | 28.8 ± 6. 9 | 20.7 ± 6.8 | 20.7 ± 2.6 |
| Enzyme Source | Parameter | No BSA | 0.2% BSA | 0.5% BSA | 1% BSA | 2% BSA |
|---|---|---|---|---|---|---|
| UGT1A9 | Km (μM) | 17.7 ± 4.6 | 20.8 ± 4.3 | 1.61 ± 0.52 | 1.23 ± 0.42 | 1.06 ± 0.21 |
| Vmax (pmol/min/mg) | 1.79 ± 0.43 | 10.1 ± 4.5 | 6.39 ± 0.93 | 5.73 ± 0.70 | 3.85 ± 0.22 | |
| Ki (μM) | 508.8 ± 134.4 | 236.1 ± 46.1 | 251.7 ± 38.7 | 96.6 ± 36.4 | 185.2 ± 52.8 | |
| CLint (μL/min/mg) | 0.10 ± 0.03 | 0.49 ± 0.19 | 4.14 ± 0.80 | 5.52 ± 1.48 | 3.75 ± 0.92 | |
| UGT2B4 | Km (μM) | 79.3 ± 9.5 | 7.40 ± 2.40 | 1.65 ± 0.28 | 3.70 ± 0.96 | 3.65 ± 0.70 |
| Vmax (pmol/min/mg) | 13.6 ± 2.0 | 33.2 ± 3.4 | 43.8 ± 1.9 | 28.7 ± 2.2 | 21.5 ± 1.3 | |
| Ki (μM) | 682.7 ± 148.7 | 844.0 ± 101.5 | 352.6 ± 60.6 | 321.0 ± 106.3 | 1104.0 ± 236.5 | |
| CLint (μL/min/mg) | 0.17 ± 0.04 | 4.95 ± 1.18 | 27.1 ± 5.1 | 8.20 ± 2.71 | 6.09 ± 1.55 | |
| UGT2B7 | Km (μM) | 21.6 ± 2.3 | 6.00 ± 1.31 | 2.51 ± 0.65 | 0.99 ± 0.30 | 0.56 ± 0.11 |
| Vmax (pmol/min/mg) | 4.07 ± 0.50 | 9.63 ± 1.81 | 7.10 ± 1.04 | 5.95 ± 0.72 | 4.31 ± 0.20 | |
| Ki (μM) | 478.4 ± 169.3 | 304.0 ± 153.9 | 415.6 ± 83.6 | 172.0 ± 56.5 | 146.6 ± 31.4 | |
| CLint (μL/min/mg) | 0.20 ± 0.04 | 1.52 ± 0.18 | 2.66 ± 0.68 | 6.25 ± 1.22 | 8.92 ± 0.78 | |
| UGT1A3 | Km (μM) | 37.6 ± 10.6 | 4.35 ± 1.37 | 1.40 ± 0.21 | 0.70 ± 0.16 | 0.75 ± 0.10 |
| Vmax (pmol/min/mg) | 146.8 ± 21.2 | 124.3 ± 13.5 | 107.8 ± 6.5 | 100.1 ± 14.5 | 87.4 ± 2.7 | |
| Ki (μM) | 862.7 ± 265.8 | 607.4 ± 44.6 | 499.6 ± 85.9 | 452.7 ± 154.3 | 816.5 ± 189.0 | |
| CLint (μL/min/mg) | 4.19 ± 1.21 | 30.2 ± 7.8 | 77.9 ± 9.6 | 144.9 ± 22.7 | 118.3 ± 19.5 | |
| UGT1A8 | Km (μM) | 25.6 ± 4.3 | 16.0 ± 5.7 | 4.73 ± 1.05 | 2.13 ± 0.34 | 1.29 ± 0.29 |
| Vmax (pmol/min/mg) | 24.3 ± 3.5 | 21.9 ± 1.2 | 19.3 ± 4.9 | 18.4 ± 3.6 | 17.8 ± 1.1 | |
| Ki (μM) | 606.4 ± 156.8 | 357.5 ± 91.1 | 209.6 ± 15.2 | 218.4 ± 83.7 | 179.9 ± 43.1 | |
| CLint (μL/min/mg) | 0.96 ± 0.14 | 1.52 ± 0.65 | 4.16 ± 1.10 | 8.68 ± 1.48 | 14.21 ± 2.74 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, Y.; Zhang, X.; Zhu, Y.; Wu, H.; Gu, S.; Chang, Q.; Zhou, Y.; Wang, G.; Sun, J. Atypical Kinetics and Albumin Effect of Glucuronidation of 5-n-Butyl-4-{4-[2-(1H-tetrazole-5- yl)-1H-pyrrol-1-yl]phenylmethyl}-2,4-dihydro-2-(2,6- dichlorophenyl)-3H-1,2,4-triazol-3-one, a Novel Nonpeptide Angiotensin Type 1 Receptor Antagonist, in Liver Microsomes and UDP-Glucuronosyl-transferase. Molecules 2018, 23, 688. https://doi.org/10.3390/molecules23030688
Peng Y, Zhang X, Zhu Y, Wu H, Gu S, Chang Q, Zhou Y, Wang G, Sun J. Atypical Kinetics and Albumin Effect of Glucuronidation of 5-n-Butyl-4-{4-[2-(1H-tetrazole-5- yl)-1H-pyrrol-1-yl]phenylmethyl}-2,4-dihydro-2-(2,6- dichlorophenyl)-3H-1,2,4-triazol-3-one, a Novel Nonpeptide Angiotensin Type 1 Receptor Antagonist, in Liver Microsomes and UDP-Glucuronosyl-transferase. Molecules. 2018; 23(3):688. https://doi.org/10.3390/molecules23030688
Chicago/Turabian StylePeng, Ying, Xueyuan Zhang, Yinci Zhu, Hui Wu, Shiyin Gu, Qingqing Chang, Yi Zhou, Guangji Wang, and Jianguo Sun. 2018. "Atypical Kinetics and Albumin Effect of Glucuronidation of 5-n-Butyl-4-{4-[2-(1H-tetrazole-5- yl)-1H-pyrrol-1-yl]phenylmethyl}-2,4-dihydro-2-(2,6- dichlorophenyl)-3H-1,2,4-triazol-3-one, a Novel Nonpeptide Angiotensin Type 1 Receptor Antagonist, in Liver Microsomes and UDP-Glucuronosyl-transferase" Molecules 23, no. 3: 688. https://doi.org/10.3390/molecules23030688