Identification of a Heat-Inducible Element of Cysteine Desulfurase Gene Promoter in Lentinula edodes
Abstract
:1. Introduction
2. Results
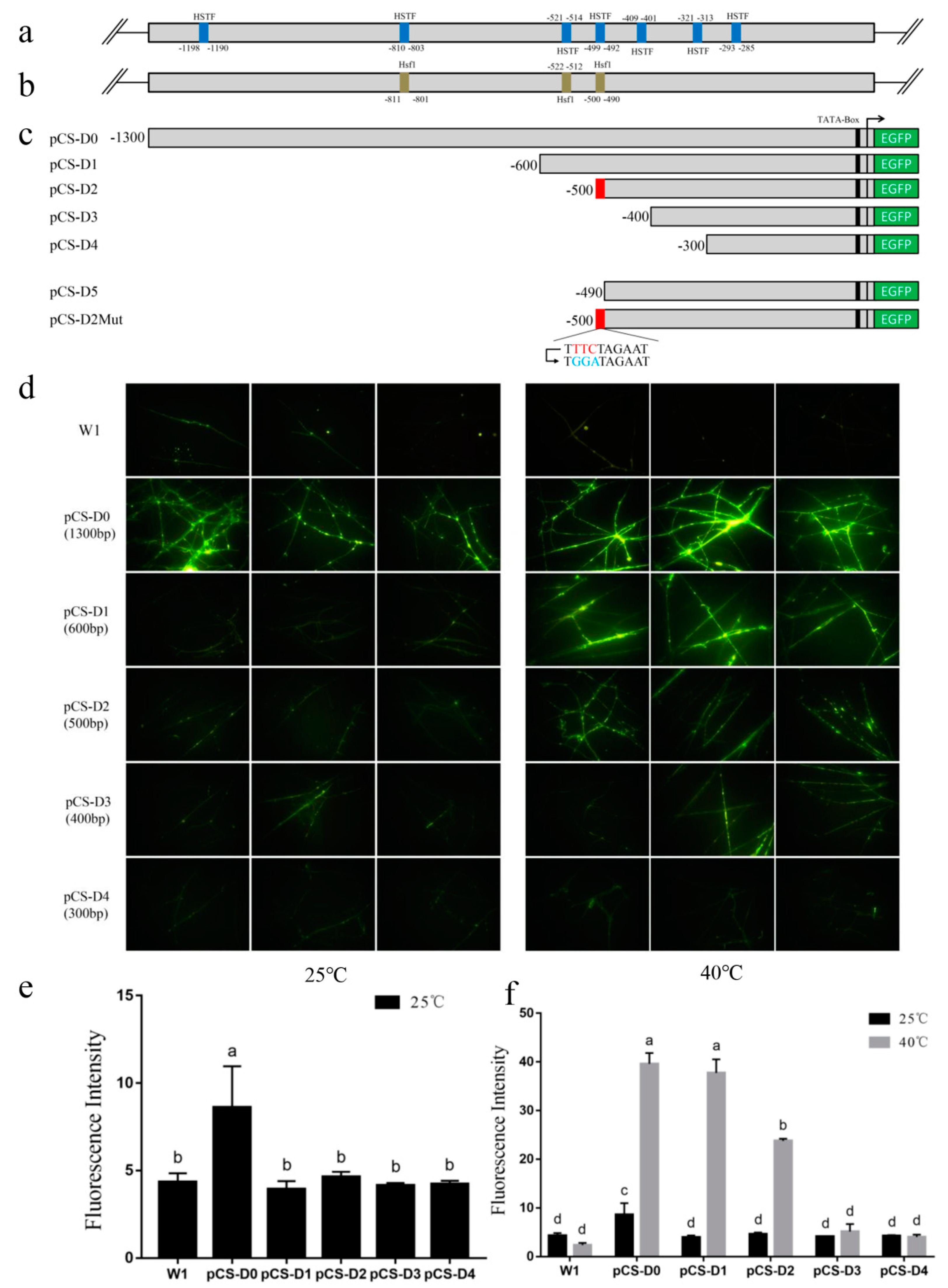
2.1. Bioinformatic Analysis of Putative Cis-Elements in pCS
2.2. EGFP Expression in Transformants Driven by Full-Length pCS
2.3. Deletion Analysis of the pCS.
2.4. Site-Directed Mutation Analysis
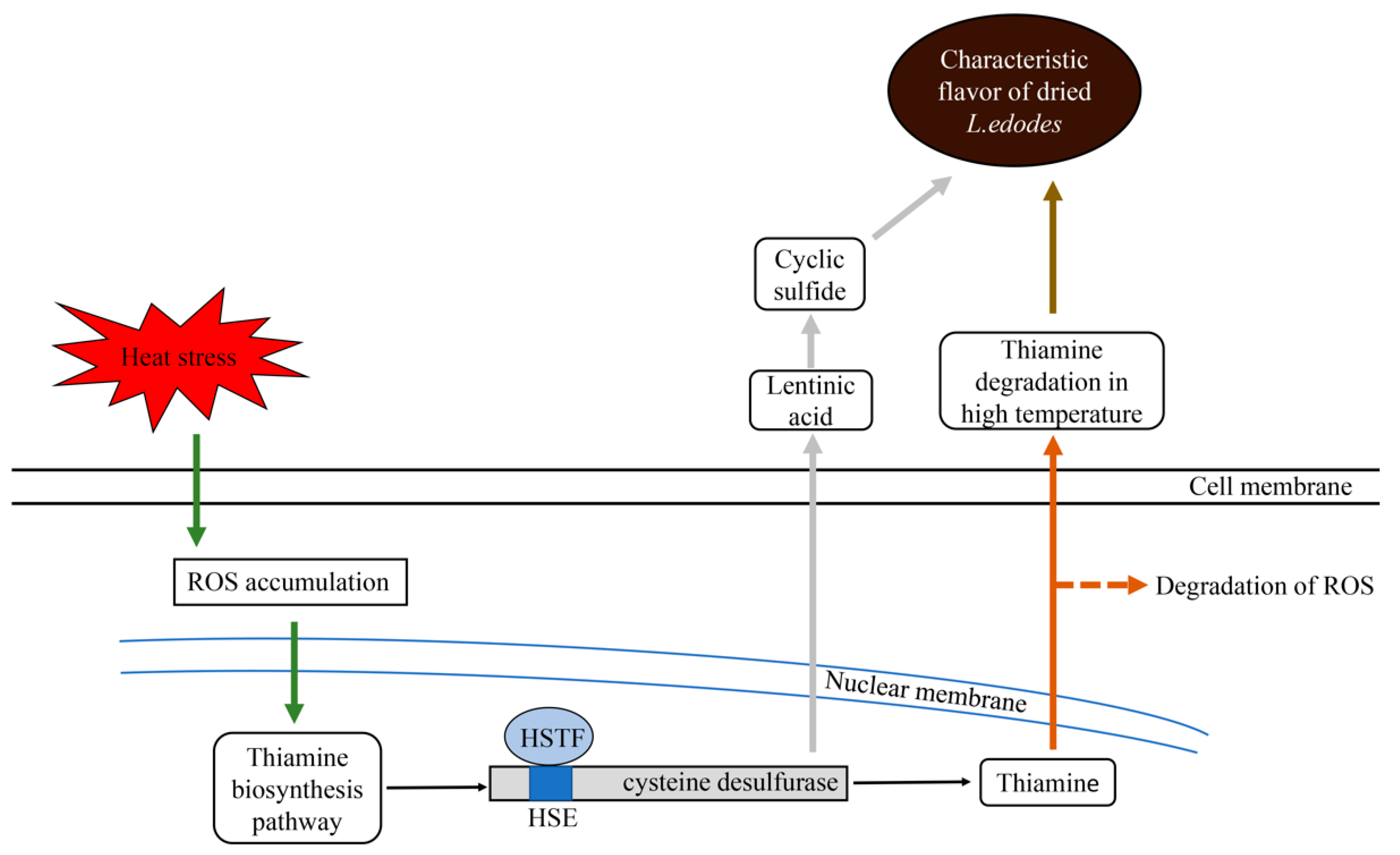
3. Discussion
4. Materials and Methods
4.1. Strains and Culture Conditions
4.2. Extraction of Genomic DNA
4.3. Cloning and Analysis of Full-Length pCS
4.4. Vector Construction of the pCS and Its Deletion Derivatives
4.5. Site-Directed Mutation and Further Truncation
4.6. Agrobacterium-Mediated Fungal Transformation
4.7. Fluorescence Microscopy and Quantification of EGFP Fluorescence Intensity
Author Contributions
Funding
Conflicts of Interest
References
- Liu, Y.; Yuan, Y.; Lei, X.; Yang, H.; Ibrahim, S.A.; Huang, W. Purification and characterisation of two enzymes related to endogenous formaldehyde in Lentinula edodes. Food Chem. 2013, 138, 2174–2179. [Google Scholar] [CrossRef] [PubMed]
- Hiraide, M.; Yokoyama, I.; Miyazaki, Y. The smell and odorous components of dried shiitake mushroom, Lentinula edodes II: Sensory evaluation by ordinary people. J. Wood Sci. 2005, 51, 628–633. [Google Scholar] [CrossRef]
- Chen, C.; Ho, C. Identification of sulfurous compounds of Shiitake mushroom (Lentinus edodes Sing.). J. Agric. Food Chem. 1986, 34, 830–833. [Google Scholar] [CrossRef]
- Politowicz, J.; Lech, K.; Lipan, L.; Figiel, A.; Carbonell-Barrachina, Á.A. Volatile composition and sensory profile of shiitake mushrooms as affected by drying method. J. Sci. Food Agric. 2018, 98, 1511–1521. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ho, C. High-performance liquid chromatographic determination of cyclic sulfur compounds of Shiitake mushroom (Lentinus edodes Sing.). J. Chrom. 1986, 356, 455–459. [Google Scholar] [CrossRef]
- Hiraide, M.; Miyazaki, Y.; Shibata, Y. The smell and odorous components of dried shiitake mushroom, Lentinula edodes I: Relationship between sensory evaluations and amounts of odorous components. J. Wood Sci. 2004, 50, 358–364. [Google Scholar] [CrossRef]
- Tian, Y.; Zhao, Y.; Huang, J.; Zeng, H.; Zheng, B. Effects of different drying methods on the product quality and volatile compounds of whole shiitake mushrooms. Food Chem. 2016, 197, 714–722. [Google Scholar] [CrossRef] [PubMed]
- Yasumoto, K.; Iwami, K.; Mitsuda, H. Enzyme-catalized evolution of Lenthionine from Lentinic Acid. Agric. Biol. Chem. 1971, 35, 2070–2080. [Google Scholar] [CrossRef]
- Yasumoto, K.; Iwami, K.; Mitsuda, H. Enzymic formation of Shiitake aroma from non-volatile precursor(s)-lenthionine from lentinic acid. Mushroom Sci. 1976, 9, 371–383. [Google Scholar]
- Hiraide, M.; Kato, A.; Nakashima, T. The smell and odorous components of dried shiitake mushroom, Lentinula edodes V: Changes in lenthionine and lentinic acid contents during the drying process. J. Wood Sci. 2010, 56, 477–482. [Google Scholar] [CrossRef]
- Liu, Y.; Lei, X.Y.; Chen, L.F.; Bian, Y.B.; Yang, H.; Ibrahim, S.A.; Huang, W. A novel cysteine desulfurase influencing organosulfur compounds in Lentinula edodes. Sci. Rep. 2015, 5, 10047. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y. Studies of the purification, cloning and biological function of key enzyme c-s lyase in the metabolism of endogenous formaldehyde in Lentinula edodes. Ph.D. Thesis, Huazhong Agricultural University, Huazhong, China, April 2014. [Google Scholar]
- Gao, S.; Wang, G.; Huang, Z.; Lei, X.; Bian, Y.; Liu, Y.; Huang, W. Selection of reference genes for qRT-PCR analysis in Lentinula edodes after hot-air drying. Molecules 2018, 24, 136. [Google Scholar] [CrossRef] [PubMed]
- Storozhenko, S.; De Pauw, P.; Van Montagu, M.; Inzé, D.; Kushnir, S. The heat-shock element Is a functional component of the Arabidopsis APX1 gene promoter. Plant Physiol. 1998, 118, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Perisic, O.; Lis, J.T. Cooperative binding of drosophila heat shock factor to arrays of a conserved 5 bp unit. Cell 1991, 64, 585–593. [Google Scholar] [CrossRef]
- Lockheart, A.; Curran, B.; Hirst, K.; Davies, M.W.; Piper, P.W.; Kingsman, A.J.; Stanway, C.A.; Ogden, J.E.; Kingsman, S.M. A heat shock element in the phosphoglycerate kinase gene promoter of yeast. Nucleic Acids Res. 1988, 16, 1333–1348. [Google Scholar] [Green Version]
- ZUBER, U.; Schumann, W. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J. Bacteriol. 1994, 176, 1359–1363. [Google Scholar] [CrossRef] [PubMed]
- Uemura, H.; Koshio, M.; Inoue, Y.; Lopez, M.C.; Baker, H.V. The Role of Gcr1p in the Transcriptional Activation of Glycolytic Genes in Yeast Saccharomyces cerevisiae. Genetics 1997, 147, 521–532. [Google Scholar]
- Peter, S.; Ayfer, B.; Marianne, K.; Thomas, N.; Klaus, S.; Joachim, M. Tec1p-independent activation of a hypha-associated Candida albicans virulence gene during infection. Infect. Immun. 2004, 72, 2386–2389. [Google Scholar]
- Yin, C.; Zhu, J.; Ma, A.; Zheng, L.; Chen, L. Characterization of the highly active fragment of glyceraldehyde-3-phosphate dehydrogenase gene promoter for recombinant protein expression in Pleurotus ostreatus. Fems Microbiol. Lett. 2015, 362, fnv010. [Google Scholar] [CrossRef]
- Cheng, S.; Yang, P.; Guo, L.; Lin, J.; Lou, N. Expression of multi-functional cellulase gene mfc in Coprinus cinereus under control of different basidiomycete promoters. Bioresour Technol. 2009, 100, 4475–4480. [Google Scholar] [CrossRef]
- Huang, X.; Lu, X.; Li, J.J. Cloning, characterization and application of a glyceraldehyde-3-phosphate dehydrogenase promoter from Aspergillus terreus. J. Ind. Microbiol. Biotechnol. 2014, 41, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Pederson, D.S. A distal heat shock element promotes the rapid response to heat shock of the HSP26 gene in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1993, 268, 7442–7448. [Google Scholar] [PubMed]
- Wang, G.; Ma, C.; Luo, Y.; Zhou, S.; Zhou, Y.; Ma, X.; Cai, Y.; Yu, J.; Bian, Y.; Gong, Y. Proteome and transcriptome reveal involvement of heat shock proteins and indoleacetic acid metabolism process in Lentinula Edodes thermotolerance. Cell. Physiol. Biochem. 2018, 50, 1617–1637. [Google Scholar] [CrossRef] [PubMed]
- Kowalska, E.; Kujda, M.; Wolak, N.; Kozik, A. Altered expression and activities of enzymes involved in thiamine diphosphate biosynthesis in Saccharomyces cerevisiae under oxidative and osmotic stress. FEMS Yeast Res. 2012, 12, 534–546. [Google Scholar] [CrossRef] [PubMed]
- Medina-Silva, R.; Barros, M.P.; Galhardo, R.S.; Netto, L.E.S.; Colepicolo, P.; Menck, C.F.M. Heat stress promotes mitochondrial instability and oxidative responses in yeast deficient in thiazole biosynthesis. Res. Microbiol. 2006, 157, 275–281. [Google Scholar] [CrossRef]
- Arnold, R.G.; Libbey, L.M.; Lindsay, R.C. Volatile flavor compounds produced by heat degradation of thiamine (vitamin B1). J. Agric. Food Chem. 1969, 17, 390–392. [Google Scholar] [CrossRef]
- Monnerjahn, C.; Techel, D.; Meyer, U.; Rensing, L. Interaction of the Neurospora crassa heat shock factor with the heat shock element during heat shock and different developmental stages. Fems Microbiol. Lett. 2000, 185, 255–261. [Google Scholar]
- Gross, D.S.; English, K.E.; Collins, K.W.; Lee, S.W. Genomic footprinting of the yeast HSP82 promoter reveals marked distortion of the DNA helix and constitutive occupancy of heat shock and TATA elements. J. Mol. Biol. 1990, 216, 611–631. [Google Scholar] [CrossRef]
- Yang, W.M.; Gahl, W.; Hamer, D. Role of heat shock transcription factor in yeast metallothionein gene expression. Mol. Cell. Biol. 1991, 11, 3676–3681. [Google Scholar] [CrossRef]
- Zhong, T.; Luke, M.T.; Arndt, K. Transcriptional regulation of the Yeast DnaJ homologue SIS1. J. Biol. Chem. 1996, 271, 1349–1356. [Google Scholar] [CrossRef]
- J. Bonner, J.; Ballou, C.; Fackenthal, D. Interactions between DNA-bound trimers of the yeast heat shock factor. Mol. Cell. Biol. 1994, 14, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Young, M.R.; Craig, E.A. Saccharomyces cerevisiae HSP70 heat shock elements are functionally distinct. Mol. Cell. Biol. 1993, 13, 5637–5646. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhou, S.; Luo, Y.; Ma, C.; Gong, Y.; Zhou, Y.; Gao, S.; Huang, Z.; Yan, L.; Hu, Y.; et al. The heat shock protein 40 LeDnaJ regulates stress resistance and indole-3-acetic acid biosynthesis in Lentinula edodes. Fungal Genet. Biol. 2018, 118, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Gong, Y.; Cai, Y.; Liu, W.; Zhou, Y.; Xiao, Y.; Xu, Z.; Liu, Y.; Lei, X.; Wang, G. Genome Sequence of the Edible Cultivated Mushroom Lentinula edodes (Shiitake) reveals insights into lignocellulose degradation. PLoS ONE 2016, 11, e0160336. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the constructs are available from the authors. |


| Element Name | Signal Sequence | Putative Function | Numbers |
|---|---|---|---|
| GCN4 | TGAATA | Transcriptional activator binding site | 17 |
| HSTF | TTCAACGAA | Involved in heat response | 7 |
| REB1 | CCACCCG | RNA polymerase IIbinding site | 1 |
| ECB | GGAAAAA | Early cell-cycle box element | 1 |
| ADR1 | TCTCC | Transcriptional activator binding site | 3 |
| GCR1 | CTTCC | Involved in decomposition of sugar | 7 |
| CuRE | GAGCAAA | Cu2+ response element | 1 |
| PHO4 | CACGTT | Activation of phosphate metabolism related genes | 2 |
| ABF1 | TCATTCCAGACG | Transcriptional activation of numerous genes | 1 |
| TBP | TATATA | TATA-Box binding protein | 2 |
| UASPHR | CTTCCT | Involved in DNA repair | 1 |
| STRE | AGGGG | Involved in stress response | 1 |
| Element Name | Signal Sequence | Putative Function | Numbers |
|---|---|---|---|
| Ash1 | YTGAT | Transcriptional inhibition of HO gene | 9 |
| Cat8, Sip4 | NCCDTYNVNCCNG | Involved in the rearrangement of carbon metabolism | 1 |
| Crz1 | GNGGCKCA | Involved in calcineurin activation | 1 |
| Fkh1, Fkh2 | RYMAAYA | Involved in cell cycle and differentiation | 3 |
| Gat1, Gln3, Gzf3 | GATAAG | Involved in glyceride metabolism | 1 |
| Gcn4 | TGATTCA | Activating amino acid synthesis related genes | 1 |
| Gcr1 | CTTCC/CWTCC | Regulation of glycolytic related genes | 9 |
| Gis1, Msn2, Msn4, Rph1, YER130C | AGGGG | Regulation of diphosphate pyrophosphate metabolism | 1 |
| Hsf1 | NTTCNNGAAN | HSTF binding site | 6 |
| Mac1 | TTTGCKCR | Cu2+ response element | 1 |
| Mot3 | AAGGWT | Involved in oxygen stress | 8 |
| Msn2, Msn4, Rph1 | CCCTC | Involved in stress response | 1 |
| Nrg1 | CCCTC | Regulates glucose metabolism and response to alkali | 2 |
| Pho4 | CACGTK | Response to phosphate limitation | 2 |
| Rgt1 | CGGANNA | Regulation of multiple glucose transporter genes | 1 |
| Rtg1, Rtg3 | GTCAC/GGTAC | Involved in interorganelle communication | 2 |
| Skn7 | GGCCAGA | Response to oxidative stress and osmoregulation | 1 |
| Stb5 | CGGNS | Regulating multidrug resistance and oxidative stress response | 15 |
| Tec1 | CATTCT | Regulating hyphal growth | 2 |
| Xbp1 | CTCGA | Cyclin gene transcriptional repression | 2 |
| Yap1 | TGACAA | Required for oxidative stress | 2 |
| Rim101 | TGCCAAG | Response to pH and in cell wall construction | 2 |
| Haa1 | SMGGSG | Involved in adaptation to weak acid stress | 3 |
| Com2 | ATAGGGT | Involved in adaptation to stress | 1 |
| Primer Name | Primer Sequence(5′-3′) |
|---|---|
| pCS-R | ccttgctcaccatGTTCAGTTAATCAAGGGGGTGAGG |
| pCSD0-F | tctagaggatccccgggtaccATGGGTGAATATAGAGAGGCGG |
| pCSD1-F | tctagaggatccccgggtaccCTGTAGCAGATTCTGAAAAGATTGTAGC |
| pCSD2-F | tctagaggatccccgggtaccTTTCTAGAATCAGTTTGATTCAGGTCTG |
| pCSD3-F | tctagaggatccccgggtaccTGAGATCTCATGCTACAGTGTGCA |
| pCSD4-F | tctagaggatccccgggtaccAGGTAAGGAACTGTCCTTGATTTCA |
| pCSD5-F | tctagaggatccccgggtaccCAGTTTGATTCAGGTCTGATTCGG |
| EGFP-F | actgaacATGGTGAGCAAGGGCGAGG |
| EGFP-R | ccacctcaaacttcggaattcTTACTTGTACAGCTCGTCCATGCC |
| hph-F | TCGTCCATCACAGTTTGCC |
| hph-R | TGCCTCTAATCCCTTGCTC |
| qEGFP-F | AAGGGCATCGACTTCAAGGAG |
| qEGFP-R | GTTCACCTTGATGCCGTTCTTC |
| pCSD2Mut-F | GCCGAATTCTGGATAGAATCAGTTTGATT |
| pCSD2Mut-R | GGTACCTTACTTGTACAGCTCGTCCAT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Z.; Lei, X.; Feng, X.; Gao, S.; Wang, G.; Bian, Y.; Huang, W.; Liu, Y. Identification of a Heat-Inducible Element of Cysteine Desulfurase Gene Promoter in Lentinula edodes. Molecules 2019, 24, 2223. https://doi.org/10.3390/molecules24122223
Huang Z, Lei X, Feng X, Gao S, Wang G, Bian Y, Huang W, Liu Y. Identification of a Heat-Inducible Element of Cysteine Desulfurase Gene Promoter in Lentinula edodes. Molecules. 2019; 24(12):2223. https://doi.org/10.3390/molecules24122223
Chicago/Turabian StyleHuang, Zhicheng, Xiaoyu Lei, Xi Feng, Shuangshuang Gao, Gangzheng Wang, Yinbing Bian, Wen Huang, and Ying Liu. 2019. "Identification of a Heat-Inducible Element of Cysteine Desulfurase Gene Promoter in Lentinula edodes" Molecules 24, no. 12: 2223. https://doi.org/10.3390/molecules24122223





