Structure and Function of Multimeric G-Quadruplexes
Abstract
1. Introduction
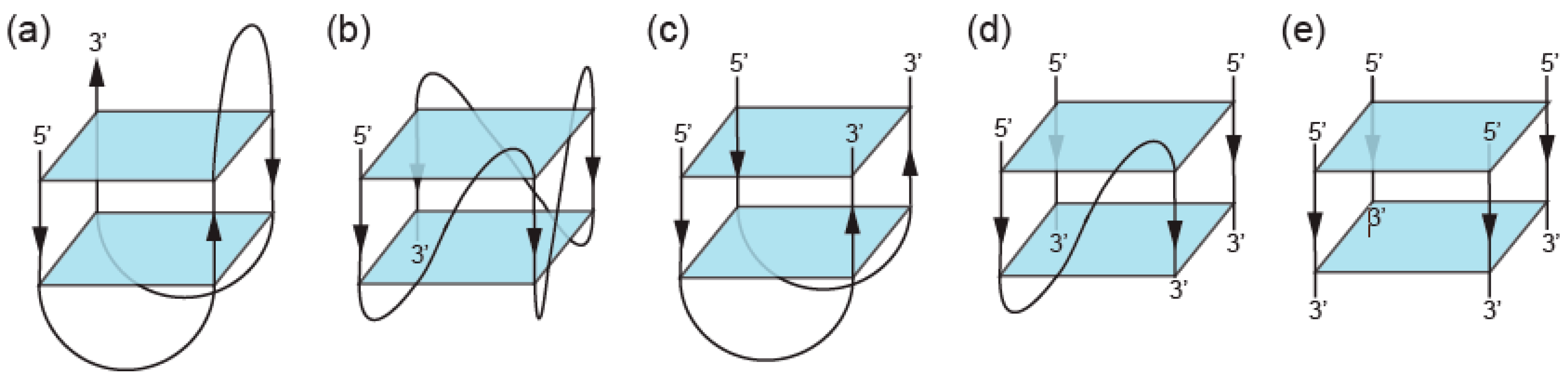
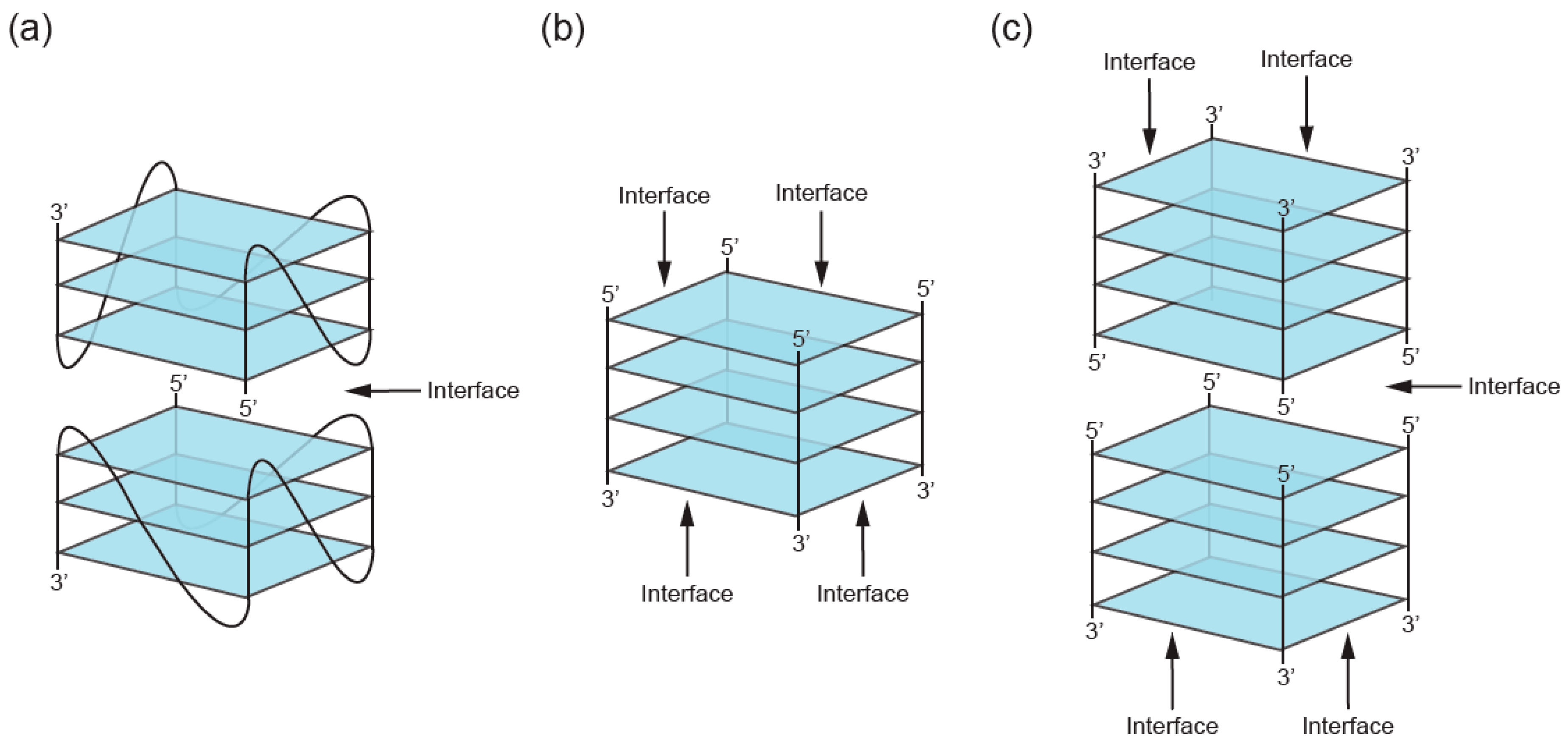
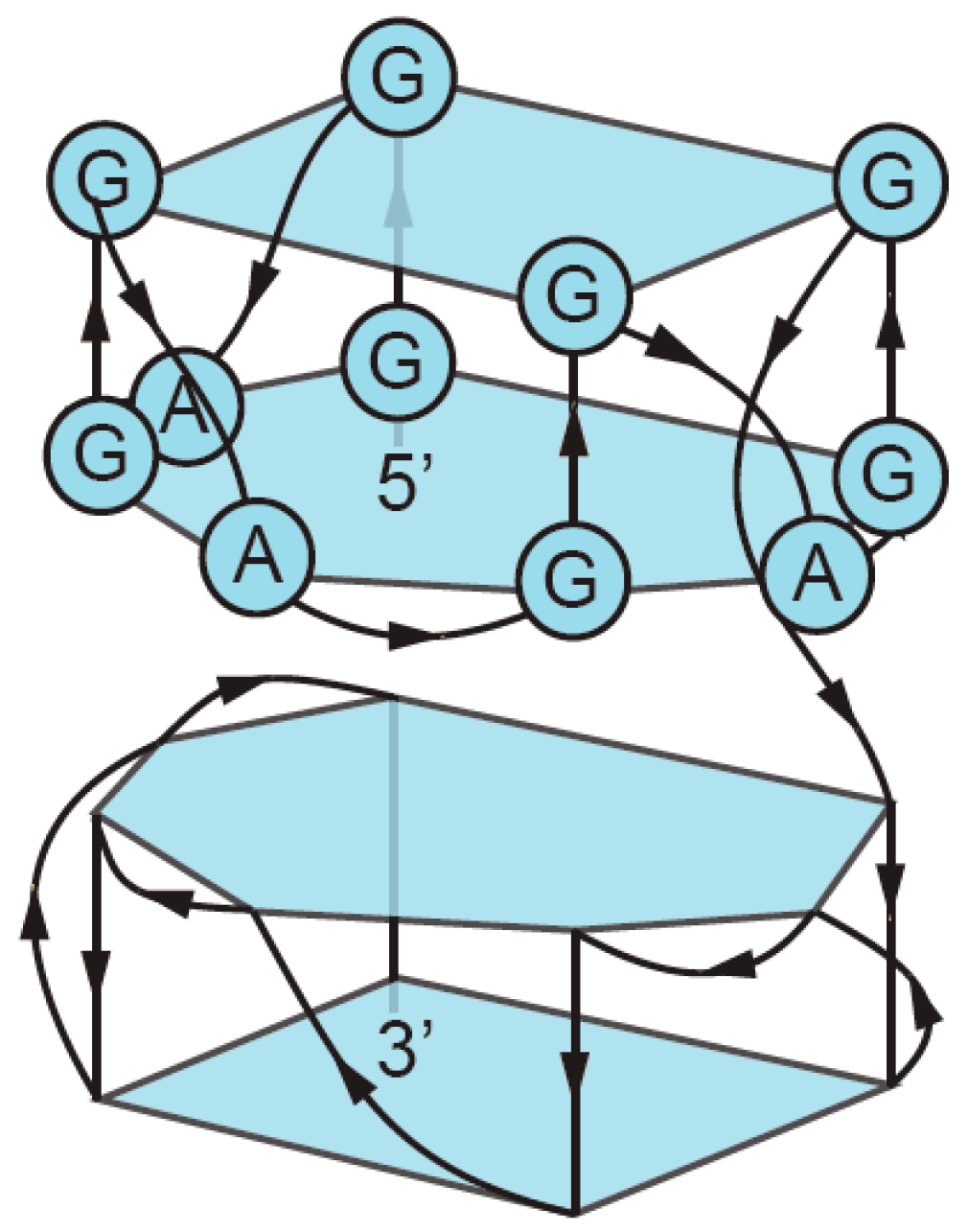
2. Definition of Multimeric G-Quadruplexes and Modes of Multimerization
3. Sequence Requirements of Multimeric G-Quadruplexes
4. Potential Biological Roles of Multimeric G-Quadruplexes at the Tips of Telomeres
5. Potential Biological Roles of Multimeric G-Quadruplexes in Promoters
6. Potential Biological Roles of RNA–DNA Hybrid G-Quadruplexes
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rich, A. DNA comes in many forms. Gene 1993, 135, 99–109. [Google Scholar] [CrossRef]
- Hu, Y.; Cecconello, A.; Idili, A.; Ricci, F.; Willner, I. Triplex DNA nanostructures: From basic properties to applications. Angew. Chem. Int. Ed. Engl. 2017, 56, 15210–15233. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.T. G-quartets 40 years later: From 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 2004, 43, 668–698. [Google Scholar] [CrossRef] [PubMed]
- Day, H.A.; Pavlou, P.; Waller, Z.A. I-Motif DNA: Structure, stability and targeting with ligands. Bioorg. Med. Chem. 2014, 22, 4407–4418. [Google Scholar] [CrossRef] [PubMed]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T. Human telomeric G-quadruplex: Structures of DNA and RNA sequences. FEBS J. 2010, 277, 1107–1117. [Google Scholar] [CrossRef] [PubMed]
- Yatsunyk, L.A.; Mendoza, O.; Mergny, J.L. “Nano-oddities”: Unusual nucleic acid assemblies for DNA-based nanostruc-tures and nanodevices. Acc. Chem. Res. 2014, 47, 1836–1844. [Google Scholar] [CrossRef] [PubMed]
- Mergny, J.L.; Sen, D. DNA quadruple helices in nano-technology. Chem. Rev. 2019, 119, 6290–6325. [Google Scholar]
- Huppert, J.L.; Balasubramanian, S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. [Google Scholar] [CrossRef]
- Todd, A.K.; Johnston, M.; Neidle, S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005, 33, 2901–2907. [Google Scholar] [CrossRef]
- Lam, E.Y.N.; Beraldi, D.; Tannahill, D.; Balasubramanian, S. G-quadruplex structures are stable and detectable in human genomic DNA. Nat. Commun. 2013, 4, 1796. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef]
- Huppert, J.L.; Balasubramanian, S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007, 35, 406–413. [Google Scholar] [CrossRef]
- Kendrick, S.; Hurley, L.H. The role of G-quadruplex/i-motif secondary structures as cis-acting regulatory elements. Pure Appl. Chem. 2010, 82, 1609–1621. [Google Scholar] [CrossRef]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef]
- Chambers, V.S.; Marsico, G.; Boutell, J.M.; Di Antonio, M.; Smith, G.P.; Balasubramanian, S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015, 33, 877–881. [Google Scholar] [CrossRef]
- Fujioka, A.; Terai, K.; Itoh, R.E.; Aoki, K.; Nakamura, T.; Kuroda, S.; Nishida, E.; Matsuda, M. Dynamics of the Ras/ERK MAPK cascade as monitored by fluorescent probes. J. Biol. Chem. 2006, 281, 8917–8926. [Google Scholar] [CrossRef]
- Kolesnikova, S.; Hubálek, M.; Bednárová, L.; Cvačka, J.; Curtis, E.A. Multimerization rules for G-quadruplexes. Nucleic Acids Res. 2017, 45, 8684–8696. [Google Scholar] [CrossRef]
- Traut, T.W. Dissociation of enzyme oligomers: A mechanism for allosteric regulation. Crit. Rev. Biochem. Mol. Biol. 1994, 29, 125–163. [Google Scholar] [CrossRef]
- Goodsell, D.S.; Olson, A.J. Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 105–153. [Google Scholar] [CrossRef]
- Amoutzias, G.D.; Robertson, D.L.; Van de Peer, Y.; Oliver, S.G. Choose your partners: Dimerization in eukaryotic transcription factors. Trends Biochem. Sci. 2008, 33, 220–229. [Google Scholar] [CrossRef]
- Matthews, J.M.; Sunde, M. Dimers, oligomers, everywhere. Adv. Exp. Med. Biol. 2012, 747, 1–18. [Google Scholar]
- Watson, J.D.; Crick, F.H.C. Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Yakovchuk, P.; Protozanova, E.; Frank-Kamenetskii, M.D. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006, 34, 564–574. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Smirnov, I.; Shafer, R.H. Effect of loop sequence and size on DNA aptamer stability. Biochemistry 2000, 39, 1462–1468. [Google Scholar] [CrossRef]
- Bourdoncle, A.; Estevez Torres, A.; Gosse, C.; Lacroix, L.; Vekhoff, P.; Le Saux, T.; Jullien, L.; Mergny, J.L. Quadruplex-based molecular beacons as tunable DNA probes. J. Am. Chem. Soc. 2006, 128, 11094–11105. [Google Scholar] [CrossRef]
- Guedin, A.; Gros, J.; Alberti, P.; Mergny, J.L. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010, 38, 7858–7868. [Google Scholar] [CrossRef]
- Mukundan, V.T.; Phan, A.T. Bulges in G-quadruplexes: Broadening the definition of G-quadruplex-forming sequences. J. Am. Chem. Soc. 2013, 135, 5017–5028. [Google Scholar] [CrossRef]
- Cheong, C.; Moore, P.B. Solution structure of an unusually stable RNA tetraplex containing G- and U-quartet structures. Biochemistry 1992, 31, 8406–8414. [Google Scholar] [CrossRef]
- Patel, P.K.; Hosur, R.V. NMR observation of T-tetrads in a parallel stranded DNA quadruplex formed by Saccharomyces cerevisiae telomere repeats. Nucleic Acids Res. 1999, 27, 2457–2464. [Google Scholar] [CrossRef]
- Patel, P.K.; Bhavesh, N.S.; Hosur, R.V. NMR observation of a novel C-tetrad in the structure of the SV40 repeat sequence GGGCGG. Biochem. Biophys. Res. Commun. 2000, 270, 967–971. [Google Scholar] [CrossRef]
- Zhang, N.; Gorin, A.; Majumdar, A.; Kettani, A.; Chernichenko, N.; Skripkin, E.; Patel, D.J. Dimeric DNA quadruplex containing major groove-aligned A-T-A-T and G-C-G-C tetrads stabilized by inter-subunit Watson-Crick A-T and G-C pairs. J. Mol. Biol. 2001, 312, 1073–1088. [Google Scholar] [CrossRef]
- Pan, B.; Xiong, Y.; Shi, K.; Deng, J.; Sundaralingam, M. Crystal structure of an RNA purine-rich tetraplex containing adenine tetrads: Implications for specific binding in RNA tetraplexes. Structure 2003, 11, 815–823. [Google Scholar] [CrossRef]
- Kimura, T.; Xu, Y.; Komiyama, M. Human telomeric RNA r(UAGGGU) sequence forms parallel tetraplex structure with U-quartet. Nucleic Acids Symp. Ser. (Oxf.) 2009, 53, 239–240. [Google Scholar] [CrossRef]
- Virgilio, A.; Esposito, V.; Citarella, G.; Mayol, L.; Galeone, A. Structural investigations on the anti-HIV G-quadruplex-forming oligonucleotide TGGGAG and its analogues: Evidence for the presence of an A-tetrad. ChemBioChem 2012, 13, 2219–2224. [Google Scholar] [CrossRef]
- Huang, H.; Suslov, N.B.; Li, N.S.; Shelke, S.A.; Evans, M.E.; Koldobskaya, Y.; Rice, P.A.; Piccirilli, J.A. A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat. Chem. Biol. 2014, 10, 686–691. [Google Scholar] [CrossRef]
- Nasiri, A.H.; Wurm, J.P.; Immer, C.; Weickhmann, A.K.; Wöhnert, J. An intermolecular G-quadruplex as the basis for GTP recognition in the class V-GTP aptamer. RNA 2016, 22, 1750–1759. [Google Scholar] [CrossRef]
- Trachman, R.J., III; Demeshkina, N.A.; Lau, M.W.L.; Panchapakesan, S.S.S.; Jeng, S.C.Y.; Unrau, P.J.; Ferré-D′Amaré, A.R. Structural basis for high-affinity fluorophore binding and activation by RNA mango. Nat. Chem. Biol. 2017, 13, 807–813. [Google Scholar] [CrossRef]
- Chaires, J.B.; Trent, J.O.; Gray, R.D.; Dean, W.L.; Buscaglia, R.; Thomas, S.D.; Miller, D.M. An Improved model for the hTERT promoter quadruplex. PLoS ONE 2014, 9, e115580. [Google Scholar] [CrossRef]
- Do, N.Q.; Lim, K.W.; Teo, M.H.; Heddi, B.; Phan, A.T. Stacking of G-quadruplexes: NMR structure of a G-rich oligonucleotide with potential anti-HIV and anticancer activity. Nucleic Acids Res. 2011, 39, 9448–9457. [Google Scholar] [CrossRef]
- Kato, Y.; Ohyama, T.; Mita, H.; Yamamoto, Y. Dynamics and thermodynamics of dimerization of parallel G-quadruplexed DNA formed from d(TTAGn) (n = 3−5). J. Am. Chem. Soc. 2005, 127, 9980–9981. [Google Scholar] [CrossRef]
- Lech, C.J.; Heddi, B.; Phan, A.T. Guanine base stacking in G-quadruplex nucleic acids. Nucleic Acids Res. 2013, 41, 2034–2046. [Google Scholar] [CrossRef]
- Smargiasso, N.; Rosu, F.; Hsia, W.; Colson, P.; Baker, E.S.; Bowers, M.T.; De Pauw, E.; Gabelica, V. G-Quadruplex DNA assemblies: Loop length, cation identity, and multimer formation. J. Am. Chem. Soc. 2008, 130, 10208–10216. [Google Scholar] [CrossRef]
- Haider, S.M.; Parkinson, G.N.; Neidle, S. Molecular dynamics and principal components analysis of human telomeric quadruplex multimers. Biophys. J. 2008, 95, 296–311. [Google Scholar] [CrossRef]
- Petraccone, L.; Trent, J.O.; Chaires, J.B. The tail of the telomere. J. Am. Chem. Soc. 2008, 130, 16530–16532. [Google Scholar] [CrossRef]
- Monchaud, D.; Teulade-Fichou, M.P. A hitchhiker′s guide to G-quadruplex ligands. Org. Biomol. Chem. 2008, 6, 627–636. [Google Scholar] [CrossRef]
- Bai, L.P.; Hagihara, M.; Jiang, Z.H.; Nakatani, K. Ligand binding to tandem G quadruplexes from human telomeric DNA. ChemBioChem 2008, 9, 2583–2587. [Google Scholar] [CrossRef]
- Haider, S.M.; Neidle, S.; Parkinson, G.N. A structural analysis of G-quadruplex/ligand interactions. Biochimie 2011, 93, 1239–1251. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, Y.; Zhang, W. G-quadruplex structures and their interaction diversity with ligands. ChemMedChem 2014, 9, 899–911. [Google Scholar] [CrossRef]
- Hu, M.H.; Chen, S.B.; Wang, B.; Ou, T.M.; Gu, L.Q.; Tan, J.H.; Huang, Z.S. Specific targeting of telomeric multimeric G-quadruplexes by a new triaryl-substituted imidazole. Nucleic Acids Res. 2017, 45, 1606–1618. [Google Scholar] [CrossRef]
- Funke, A.; Karg, B.; Dickerhoff, J.; Balke, D.; Müller, S.; Weisz, K. Ligand- induced dimerization of a truncated parallel MYC G-Quadruplex. ChemBioChem 2018, 19, 505–512. [Google Scholar] [CrossRef]
- Krishnan-Ghosh, Y.; Liu, D.; Balasubramanian, S. Formation of an interlocked quadruplex dimer by d(GGGT). J. Am. Chem. Soc. 2004, 126, 11009–11016. [Google Scholar] [CrossRef]
- Sen, D.; Gilbert, W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 1988, 334, 65–70. [Google Scholar] [CrossRef]
- Sundquist, W.I.; Klug, A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature 1989, 342, 825–829. [Google Scholar] [CrossRef]
- Williamson, J.R.; Raghuraman, M.K.; Cech, T.R. Monovalent cation-induced structure of telomeric DNA: The G-quartet model. Cell 1989, 59, 871–880. [Google Scholar] [CrossRef]
- Sen, D.; Gilbert, W. Novel DNA superstructures formed by telomere-like oligomers. Biochemistry 1992, 31, 65–70. [Google Scholar] [CrossRef]
- Laughlan, G.; Murchie, A.I.; Norman, D.G.; Moore, M.H.; Moody, P.C.; Lilley, D.M.; Luisi, B. The high-resolution crystal structure of a parallel-stranded guanine tetraplex. Science 1994, 265, 520–524. [Google Scholar] [CrossRef]
- Phillips, K.; Dauter, Z.; Murchie, A.I.; Lilley, D.M.; Luisi, B. The crystal structure of a parallel-stranded guanine tetraplex at 0.95 A resolution. J. Mol. Biol. 1997, 273, 171–182. [Google Scholar] [CrossRef]
- Borbone, N.; Amato, J.; Oliviero, G.; D′Atri, V.; Gabelica, V.; De Pauw, E.; Piccialli, G.; Mayol, L. d(CGGTGGT) forms an octameric parallel G-quadruplex via stacking of unusual G(:C):G(:C):G(:C):G(:C) octads. Nucleic Acids Res. 2011, 39, 7848–7857. [Google Scholar] [CrossRef]
- D′Atri, V.; Borbone, N.; Amato, J.; Gabelica, V.; D′Errico, S.; Piccialli, G.; Mayol, L.; Oliviero, G. DNA-based nanostructures: The effect of the base sequence on octamer formation from d(XGGYGGT) tetramolecular G-quadruplexes. Biochimie 2014, 99, 119–128. [Google Scholar] [CrossRef]
- Kuryavyi, V.; Cahoon, L.A.; Seifert, H.S.; Patel, D.J. RecA-binding pilE G4 sequence essential for pilin antigenic variation forms monomeric and 5′ end-stacked dimeric parallel G-quadruplexes. Structure 2012, 20, 2090–2102. [Google Scholar] [CrossRef]
- Adrian, M.; Ang, D.J.; Lech, C.J.; Heddi, B.; Nicolas, A.; Phan, A.T. Structure and conformational dynamics of a stacked dimeric G-quadruplex formed by the human CEB1 minisatellite. J. Am. Chem. Soc. 2014, 136, 6297–6305. [Google Scholar] [CrossRef]
- Matsugami, A.; Ouhashi, K.; Kanagawa, M.; Liu, H.; Kanagawa, S.; Uesugi, S.; Katahira, M. An intramolecular quadruplex of (GGA) (4) triplet repeat DNA with a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad, and its dimeric interaction. J. Mol. Biol. 2001, 313, 255–269. [Google Scholar] [CrossRef]
- Kuryavyi, V.; Phan, A.T.; Patel, D.J. Solution structures of all parallel-stranded monomeric and dimeric G-quadruplex scaffolds of the human c-kit2 promoter. Nucleic Acids Res. 2010, 38, 6757–6773. [Google Scholar] [CrossRef]
- Trajkovski, M.; da Silva, M.W.; Plavec, J. Unique structural features of interconverting monomeric and dimeric G-quadruplexes adopted by a sequence from the intron of the N-myc gene. J. Am. Chem. Soc. 2012, 134, 4132–4141. [Google Scholar] [CrossRef]
- Wei, D.; Todd, A.K.; Zloh, M.; Gunaratnam, M.; Parkinson, G.N.; Neidle, S. Crystal structure of a promoter sequence in the B-raf gene reveals an intertwined dimer quadruplex. J. Am. Chem. Soc. 2013, 135, 19319–19329. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, D. Sequence, stability, and structure of G-quadruplexes and their interactions with drugs. Curr. Protoc. Nucleic Acid Chem. 2012, 50, 1–17. [Google Scholar]
- Yaku, H.; Murashima, T.; Tateishi-Karimata, H.; Nakano, S.; Miyoshi, D.; Sugimoto, N. Study on effects of molecular crowding on G-quadruplex-ligand binding and ligand-mediated telomerase inhibition. Methods 2013, 64, 19–27. [Google Scholar] [CrossRef]
- Bhattacharyya, D.; Mirihana Arachchilage, G.; Basu, S. Metal cations in G- quadruplex folding and stability. Front. Chem. 2016, 4, 38. [Google Scholar] [CrossRef]
- Kar, A.; Jones, N.; Arat, N.Ö.; Fishel, R.; Griffith, J.D. Long repeating (TTAGGG)n single-stranded DNA self-condenses into compact beaded filaments stabilized by G-quadruplex formation. J. Biol. Chem. 2018, 293, 9473–9485. [Google Scholar] [CrossRef]
- Podbevsek, P.; Plavec, J. KRAS promoter oligonucleotide with decoy activity dimerizes into a unique topology consisting of two G-quadruplex units. Nucleic Acids Res. 2016, 44, 917–925. [Google Scholar] [CrossRef]
- Marsh, T.C.; Henderson, E. G-wires: Self-assembly of a telomeric oligonucleotide, d(GGGGTTGGGG), into large superstructures. Biochemistry 1994, 33, 10718–10724. [Google Scholar] [CrossRef]
- Bose, K.; Lech, C.J.; Heddi, B.; Phan, A.T. High-resolution AFM structure of DNA G-wires in aqueous solution. Nat. Commun. 2018, 9, 1959. [Google Scholar] [CrossRef]
- Hessari, N.M.; Spindler, L.; Troha, T.; Lam, W.C.; Drevensek-Olenik, I.; da Silva, M.W. Programmed self-assembly of a quadruplex DNA nanowire. Chemistry 2014, 20, 3626–3630. [Google Scholar] [CrossRef]
- Guo, X.; Liu, S.; Yu, Z. Bimolecular quadruplexes and their transitions to higher-order molecular structures detected by ESI-FTICR-MS. J. Am. Soc. Mass Spectrom. 2007, 18, 1467–1476. [Google Scholar] [CrossRef]
- Majerová, T.; Streckerová, T.; Bednárová, L.; Curtis, E.A. Sequence requirements of intrinsically fluorescent G-quadruplexes. Biochemistry 2018, 57, 4052–4062. [Google Scholar] [CrossRef]
- Kolesnikova, S.; Srb, P.; Vrzal, L.; Lawrence, M.; Veverka, V.; Curtis, E.A. GTP-dependent formation of multimeric G-quadruplexes. ACS Chem. Biol. in press. [CrossRef]
- Švehlová, K.; Lawrence, M.S.; Bednárová, L.; Curtis, E.A. Altered biochemical specificity of G-quadruplexes with mutated tetrads. Nucleic Acids Res. 2016, 44, 10789–10803. [Google Scholar] [CrossRef]
- Lu, M.; Guo, Q.; Kallenbach, N.R. Structure and stability of sodium and potassium complexes of dT4G4 and dT4G4T. Biochemistry 1992, 31, 2455–2459. [Google Scholar] [CrossRef]
- Creze, C.; Rinaldi, B.; Haser, R.; Bouvet, P.; Gouet, P. Structure of a d(TGGGGT) quadruplex crystallized in the presence of Li+ ions. Acta Crystallogr. D Biol. Crystallogr. 2007, 63, 682–688. [Google Scholar] [CrossRef]
- Deng, J.; Xiong, Y.; Sundaralingam, M. X-ray analysis of an RNA tetraplex (UGGGGU)(4) with divalent Sr(2+) ions at subatomic resolution (0.61 A). Proc. Nat. Acad. Sci. USA 2001, 98, 13665–13670. [Google Scholar] [CrossRef]
- Hazel, P.; Huppert, J.; Balasubramanian, S.; Neidle, S. Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004, 126, 16405–16415. [Google Scholar] [CrossRef]
- Risitano, A.; Fox, K.R. Stability of intramolecular DNA quadruplexes: Comparison with DNA duplexes. Biochemistry 2003, 42, 6507–6513. [Google Scholar] [CrossRef]
- Blackburn, E.H.; Gall, J.G. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J. Mol. Biol. 1978, 120, 33–53. [Google Scholar] [CrossRef]
- Jacob, N.K.; Skopp, R.; Price, C.M. G-overhang dynamics at Tetrahymena telomeres. EMBO J. 2001, 20, 4299–4308. [Google Scholar] [CrossRef]
- Moyzis, R.K.; Buckingham, J.M.; Cram, L.S.; Dani, M.; Deaven, L.L.; Jones, M.D.; Meyne, J.; Ratliff, R.L.; Wu, J.R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Nat. Acad. Sci. USA 1988, 85, 6622–6626. [Google Scholar] [CrossRef]
- Wright, W.E.; Tesmer, V.M.; Huffman, K.E.; Levene, S.D.; Shay, J.W. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997, 11, 2801–2809. [Google Scholar] [CrossRef]
- Henderson, E.; Hardin, C.C.; Walk, S.K.; Tinoco, I., Jr.; Blackburn, E.H. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine- guanine base pairs. Cell 1987, 51, 899–908. [Google Scholar] [CrossRef]
- Neidle, S.; Parkinson, G.N. The structure of telomeric DNA. Curr. Opin. Struct. Biol. 2003, 13, 275–283. [Google Scholar] [CrossRef]
- Li, J.; Correia, J.J.; Wang, L.; Trent, J.O.; Chaires, J.B. Not so crystal clear: The structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005, 33, 4649–4659. [Google Scholar] [CrossRef]
- Petraccone, L. Higher-order quadruplex structures. Top. Curr. Chem. 2013, 330, 23–46. [Google Scholar]
- Yu, H.Q.; Miyoshi, D.; Sugimoto, N. Characterization of structure and stability of long telomeric DNA G-quadruplexes. J. Am. Chem. Soc. 2006, 128, 15461–15468. [Google Scholar] [CrossRef]
- Kankia, B. Tetrahelical monomolecular architecture of DNA: A new building block for nanotechnology. J. Phys. Chem. B. 2014, 118, 6134–6140. [Google Scholar] [CrossRef]
- Kankia, B. Monomolecular tetrahelix of polyguanine with a strictly defined folding pattern. Sci. Rep. 2018, 8, 10115. [Google Scholar] [CrossRef]
- Xu, Y.; Komiyama, M. The structural studies of human telomeric DNA using AFM. Nucleic Acids Symp. Ser. (Oxf). 2007, 51, 241–242. [Google Scholar] [CrossRef][Green Version]
- Xu, X.; Ishizuka, T.; Kurabayashi, K.; Komiyama, M. Consecutive formation of G-quadruplexes in human telomeric-overhang DNA: A protective capping structure for telomere ends. Angew. Chem. Int. Ed. 2009, 48, 7833–7836. [Google Scholar] [CrossRef]
- Petraccone, L.; Spink, C.; Trent, J.O.; Garbett, N.C.; Mekmaysy, C.S.; Giancola, C.; Chaires, J.B. Structure and stability of higher-order human telomeric quadruplexes. J. Am. Chem. Soc. 2011, 133, 20951–20961. [Google Scholar] [CrossRef]
- Schaffitzel, C.; Berger, I.; Postberg, J.; Hanes, J.; Lipps, H.J.; Pluckthun, A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Nat. Acad. Sci. USA 2001, 98, 8572–8577. [Google Scholar] [CrossRef]
- Moye, A.L.; Porter, K.C.; Cohen, S.B.; Phan, T.; Zyner, K.G.; Sasaki, N.; Lovrecz, G.O.; Beck, J.L.; Bryan, T.M. Telomeric G-quadruplexes are a substrate and site of localization for human telomerase. Nat. Commun. 2015, 6, 7643. [Google Scholar] [CrossRef]
- Zhao, C.; Wu, L.; Ren, J.; Xu, Y.; Qu, X. Targeting human telomeric higher-order DNA: Dimeric G-quadruplex units serve as preferred binding site. J. Am. Chem. Soc. 2013, 135, 18786–18789. [Google Scholar] [CrossRef]
- Huang, X.X.; Zhu, L.N.; Wu, B.; Huo, Y.F.; Duan, N.N.; Kong, D.M. Two cationic porphyrin isomers showing different multimeric G-quadruplex recognition specificity against monomeric G-quadruplexes. Nucleic Acids Res. 2014, 42, 8719–8731. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, Y.C.; Kong, D.M.; Guo, D.S. Tetraphenylethene derivatives with different numbers of positively charged side arms have different multimeric G- quadruplex recognition specificity. Chemistry 2015, 21, 13253–13260. [Google Scholar] [CrossRef]
- Dai, J.; Dexheimer, T.S.; Chen, D.; Carver, M.; Ambrus, A.; Jones, R.A.; Yang, D. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 2006, 128, 1096–1098. [Google Scholar] [CrossRef]
- González, V.; Hurley, L.H. The c-MYC NHE III1: Function and regulation. Annu. Rev. Pharmacol. Toxicol. 2008, 50, 111–129. [Google Scholar] [CrossRef]
- Mathad, R.I.; Hatzakis, E.; Dai, J.; Yang, D. c-MYC promoter G-quadruplex formed at the 5′-end of NHE III1 element: Insights into biological relevance and parallel-stranded G-quadruplex stability. Nucleic Acids Res. 2011, 39, 9023–9033. [Google Scholar] [CrossRef]
- Kerkour, A.; Marquevielle, J.; Ivashchenko, S.; Yatsunyk, L.A.; Mergny, J.L.; Salgado, G.F. High-resolution three-dimensional NMR structure of the KRAS proto-oncogene promoter reveals key features of a G-quadruplex involved in transcriptional regulation. J. Biol. Chem. 2017, 292, 8082–8091. [Google Scholar] [CrossRef]
- Matsugami, A.; Okuizumi, T.; Uesugi, S.; Katahira, M. Intramolecular higher order packing of parallel quadruplexes comprising a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad of GGA triplet repeat DNA. J. Biol. Chem. 2003, 278, 28147–28153. [Google Scholar] [CrossRef]
- Palumbo, S.L.; Memmott, R.M.; Uribe, D.J.; Krotova-Khan, Y.; Hurley, J.H.; Ebbinghaus, S.W. A novel G-quadruplex-forming GGA repeat region in the c-myb promoter is a critical regulator of promoter activity. Nucleic Acids Res. 2008, 36, 1755–1769. [Google Scholar] [CrossRef]
- Broxson, C.; Beckett, J.; Tornaletti, S. Transcription arrest by a G-quadruplex forming-trinucleotide repeat sequence from the human c-myb gene. Biochemistry 2011, 50, 4162–4172. [Google Scholar] [CrossRef]
- Hirsch, M.R.; Gaugler, L.; Deagostini-Bazin, H.; Bally-Cuif, L.; Goridis, C. Identification of positive and negative regulatory elements governing cell-type-specific expression of the neural cell adhesion molecule gene. Mol. Cell. Biol. 1990, 10, 1959–1968. [Google Scholar] [CrossRef]
- Chamboredon, S.; Briggs, J.; Vial, E.; Hurault, J.; Galvagni, F.; Oliviero, S.; Bos, T.; Castellazzi, M. v-Jun downregulates the SPARC target gene by binding to the proximal promoter indirectly through Sp1/3. Oncogene 2003, 22, 4047–4061. [Google Scholar] [CrossRef]
- Morgan, R.K.; Batra, H.; Gaerig, V.C.; Hockings, J.; Brooks, T.A. Identification and characterization of a new G-quadruplex forming region within the kRAS promoter as a transcriptional regulator. Biochim. Biophys. Acta 2016, 1859, 235–245. [Google Scholar] [CrossRef]
- Islam, I.; Baba, Y.; Witarto, A.B.; Yoshida, W. G-quadruplex-forming GGA repeat region functions as a negative regulator of the Ccnb1ip1 enhancer. Biosci. Biotechnol. Biochem. 2019, 7, 1–6. [Google Scholar] [CrossRef]
- Palumbo, S.L.; Ebbinghaus, S.W.; Hurley, L.H. Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. J. Am. Chem. Soc. 2009, 131, 10878–10891. [Google Scholar] [CrossRef]
- Kang, H.J.; Cui, Y.; Yin, H.; Scheid, A.; Hendricks, W.P.D.; Schmidt, J.; Sekulic, A.; Kong, D.; Trent, J.M.; Gokhale, V.; et al. A pharmacological chaperone molecule induces cancer cell death by restoring tertiary DNA structures in mutant hTERT promoters. J. Am. Chem. Soc. 2016, 138, 13673–13692. [Google Scholar] [CrossRef]
- Micheli, E.; Martufi, M.; Cacchione, S.; De Santis, P.; Savino, M. Self-organization of G-quadruplex structures in the hTERT core promoter stabilized by polyaminic side chain perylene derivatives. Biophys. Chem. 2010, 153, 43–53. [Google Scholar] [CrossRef]
- Saha, D.; Singh, A.; Hussain, T.; Srivastava, V.; Sengupta, S.; Kar, A.; Dhapola, P.; Dhople, V.; Ummanni, R.; Chowdhury, S. Epigenetic suppression of human telomerase (hTERT) is mediated by the metastasis suppressor NME2 in a G-quadruplex-dependent fashion. J. Biol. Chem. 2017, 292, 15205–15215. [Google Scholar] [CrossRef]
- Yafe, A.; Etzioni, S.; Weisman-Shomer, P.; Fry, M. Formation and properties of hairpin and tetraplex structures of guanine-rich regulatory sequences of muscle-specific genes. Nucleic Acids Res. 2005, 33, 2887–2900. [Google Scholar] [CrossRef]
- Rigo, R.; Sissi, C. Characterization of G4-G4 crosstalk in the c-KIT promoter region. Biochemistry 2017, 56, 4309–4312. [Google Scholar] [CrossRef]
- Ducani, C.; Bernardinelli, G.; Högberg, B.; Keppler, B.K.; Terenzi, A. Interplay of three G-quadruplex units in the KIT promoter. J. Am. Chem. Soc. 2019, 141, 10205–10213. [Google Scholar] [CrossRef]
- Lavezzo, E.; Berselli, M.; Frasson, I.; Perrone, R.; Palù, G.; Brazzale, A.R.; Richter, S.N.; Toppo, S. G-quadruplex forming sequences in the genome of all known viruses: A comprehensive guide. PLoS Comput. Biol. 2018, 14, e1006675. [Google Scholar] [CrossRef]
- Frasson, I.; Nadai, M.; Richter, S.N. Conserved G-quadruplexes regulate the immediate early promoters of human Alphaherpesviruses. Molecules 2019, 24, 2375. [Google Scholar] [CrossRef]
- Xu, Y.; Kimura, T.; Komiyama, M. Human telomere RNA and DNA form an intermolecular G-quadruplex. Nucleic Acids Symp. Ser. (Oxf.) 2008, 52, 169–170. [Google Scholar] [CrossRef]
- Wanrooij, P.H.; Uhler, J.P.; Shi, Y.; Westerlund, F.; Falkenberg, M.; Gustafsson, C.M. A hybrid G-quadruplex structure formed between RNA and DNA explains the extraordinary stability of the mitochondrial R-loop. Nucleic Acids Res. 2012, 40, 10334–10344. [Google Scholar] [CrossRef]
- Zheng, K.W.; Wu, R.Y.; He, Y.D.; Xiao, S.; Zhang, J.Y.; Liu, J.Q.; Hao, Y.H.; Tan, Z. A competitive formation of DNA:RNA hybrid G-quadruplex is responsible to the mitochondrial transcription termination at the DNA replication priming site. Nucleic Acids Res. 2014, 42, 10832–10844. [Google Scholar] [CrossRef]
- Zheng, K.W.; Xiao, S.; Liu, J.Q.; Zhang, J.Y.; Hao, Y.H.; Tan, Z. Co-transcriptional formation of DNA:RNA hybrid G-quadruplex and potential function as constitutional cis element for transcription control. Nucleic Acids Res. 2013, 41, 5533–5541. [Google Scholar] [CrossRef]
- Xiao, S.; Zhang, J.Y.; Wu, J.; Wu, R.Y.; Xia, Y.; Zheng, K.W.; Hao, Y.H.; Zhou, X.; Tan, Z. Formation of DNA:RNA hybrid G-quadruplexes of two G-quartet layers in transcription: Expansion of the prevalence and diversity of G-quadruplexes in genomes. Angew. Chem. Int. Ed. Engl. 2014, 53, 13110–13114. [Google Scholar] [CrossRef]
- Xiao, S.; Zhang, J.Y.; Zheng, K.W.; Hao, Y.H.; Tan, Z. Bioinformatic analysis reveals an evolutional selection for DNA:RNA hybrid G-quadruplex structures as putative transcription regulatory elements in warm-blooded animals. Nucleic Acids Res. 2013, 41, 10379–10390. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Zheng, K.W.; Xiao, S.; Hao, Y.H.; Tan, Z. Mechanism and manipulation of DNA:RNA hybrid G-quadruplex formation in transcription of G-rich DNA. J. Am. Chem. Soc. 2014, 136, 1381–1390. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, J.Y.; Zhang, Z.Y.; Tong, T.J.; Hao, Y.H.; Tan, Z. Real-time detection reveals responsive cotranscriptional formation of persistent intramolecular DNA and intermolecular DNA:RNA hybrid G-quadruplexes stabilized by R-loop. Anal. Chem. 2017, 89, 6036–6042. [Google Scholar] [CrossRef]
- Shrestha, P.; Xiao, S.; Dhakal, S.; Tan, Z.; Mao, H. Nascent RNA transcripts facilitate the formation of G-quadruplexes. Nucleic Acids Res. 2014, 42, 7236–7246. [Google Scholar] [CrossRef]
- Curtis, E.A.; Liu, D.R. Discovery of widespread GTP-binding motifs in genomic RNA and DNA. Chem. Biol. 2013, 20, 521–532. [Google Scholar] [CrossRef]







© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kolesnikova, S.; Curtis, E.A. Structure and Function of Multimeric G-Quadruplexes. Molecules 2019, 24, 3074. https://doi.org/10.3390/molecules24173074
Kolesnikova S, Curtis EA. Structure and Function of Multimeric G-Quadruplexes. Molecules. 2019; 24(17):3074. https://doi.org/10.3390/molecules24173074
Chicago/Turabian StyleKolesnikova, Sofia, and Edward A. Curtis. 2019. "Structure and Function of Multimeric G-Quadruplexes" Molecules 24, no. 17: 3074. https://doi.org/10.3390/molecules24173074
APA StyleKolesnikova, S., & Curtis, E. A. (2019). Structure and Function of Multimeric G-Quadruplexes. Molecules, 24(17), 3074. https://doi.org/10.3390/molecules24173074




